6F4K
 
 | | Crystal structure of glutathione transferase Omega 3S from Trametes versicolor in complex with hexyl-glutathione | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular recognition of wood polyphenols by phase II detoxification enzymes of the white rot Trametes versicolor.
Sci Rep, 8, 2018
|
|
6F51
 
 | | Crystal structure of glutathione transferase Omega 3S from Trametes versicolor in complex with glutathionyl-phenylacetophenone | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, L-gamma-glutamyl-S-(2-biphenyl-4-yl-2-oxoethyl)-L-cysteinylglycine, ... | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2017-11-30 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Molecular recognition of wood polyphenols by phase II detoxification enzymes of the white rot Trametes versicolor.
Sci Rep, 8, 2018
|
|
4WCX
 
 | | Crystal structure of HydG: A maturase of the [FeFe]-hydrogenase | | Descriptor: | ALANINE, Biotin and thiamin synthesis associated, FE (III) ION, ... | | Authors: | Dinis, P.C, Harmer, J.E, Driesener, R.C, Roach, P.L. | | Deposit date: | 2014-09-05 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | X-ray crystallographic and EPR spectroscopic analysis of HydG, a maturase in [FeFe]-hydrogenase H-cluster assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3L06
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase E92V mutant complexed with carbamyl phosphate and N-succinyl-L-norvaline | | Descriptor: | N-(3-CARBOXYPROPANOYL)-L-NORVALINE, N-acetylornithine carbamoyltransferase, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER, ... | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-09 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | A single mutation in the active site swaps the substrate specificity of N-acetyl-L-ornithine transcarbamylase and N-succinyl-L-ornithine transcarbamylase.
Protein Sci., 16, 2007
|
|
1HQB
 
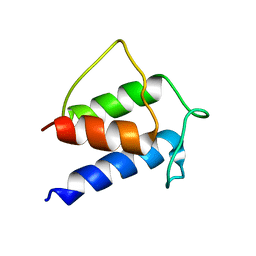 | | TERTIARY STRUCTURE OF APO-D-ALANYL CARRIER PROTEIN | | Descriptor: | APO-D-ALANYL CARRIER PROTEIN | | Authors: | Volkman, B.F, Zhang, Q, Debabov, D.V, Rivera, E, Kresheck, G, Neuhaus, F.C. | | Deposit date: | 2000-12-14 | | Release date: | 2001-08-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Biosynthesis of D-alanyl-lipoteichoic acid: the tertiary structure of apo-D-alanyl carrier protein.
Biochemistry, 40, 2001
|
|
3L05
 
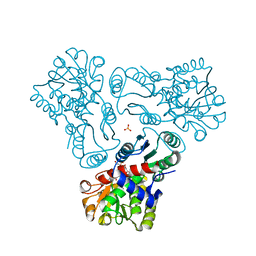 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase E92S mutant complexed with carbamyl phosphate and N-succinyl-L-norvaline | | Descriptor: | N-(3-CARBOXYPROPANOYL)-L-NORVALINE, N-acetylornithine carbamoyltransferase, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER, ... | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-09 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A single mutation in the active site swaps the substrate specificity of N-acetyl-L-ornithine transcarbamylase and N-succinyl-L-ornithine transcarbamylase.
Protein Sci., 16, 2007
|
|
1ISC
 
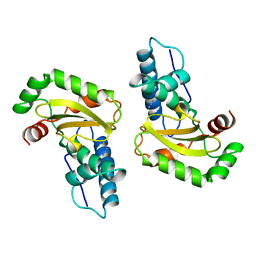 | | STRUCTURE-FUNCTION IN E. COLI IRON SUPEROXIDE DISMUTASE: COMPARISONS WITH THE MANGANESE ENZYME FROM T. THERMOPHILUS | | Descriptor: | AZIDE ION, FE (III) ION, IRON(III) SUPEROXIDE DISMUTASE | | Authors: | Lah, M.S, Dixon, M, Pattridge, K.A, Stallings, W.C, Fee, J.A, Ludwig, M.L. | | Deposit date: | 1994-07-12 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function in Escherichia coli iron superoxide dismutase: comparisons with the manganese enzyme from Thermus thermophilus.
Biochemistry, 34, 1995
|
|
6FIW
 
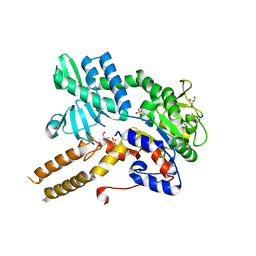 | |
6FIS
 
 | |
6F70
 
 | | Crystal structure of glutathione transferase Omega 6S from Trametes versicolor | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTATHIONE, GLYCEROL, ... | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2017-12-07 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Molecular recognition of wood polyphenols by phase II detoxification enzymes of the white rot Trametes versicolor.
Sci Rep, 8, 2018
|
|
1ISA
 
 | | STRUCTURE-FUNCTION IN E. COLI IRON SUPEROXIDE DISMUTASE: COMPARISONS WITH THE MANGANESE ENZYME FROM T. THERMOPHILUS | | Descriptor: | FE (II) ION, IRON(II) SUPEROXIDE DISMUTASE | | Authors: | Lah, M.S, Dixon, M, Pattridge, K.A, Stallings, W.C, Fee, J.A, Ludwig, M.L. | | Deposit date: | 1994-07-12 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function in Escherichia coli iron superoxide dismutase: comparisons with the manganese enzyme from Thermus thermophilus.
Biochemistry, 34, 1995
|
|
5DFE
 
 | | 70S termination complex containing E. coli RF2 | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hoffer, E.D, Dunham, C.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.09997559 Å) | | Cite: | Uniformity of Peptide Release Is Maintained by Methylation of Release Factors.
Cell Rep, 17, 2016
|
|
5CZP
 
 | | 70S termination complex containing E. coli RF2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Dunham, C.M. | | Deposit date: | 2015-07-31 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.29995918 Å) | | Cite: | Uniformity of Peptide Release Is Maintained by Methylation of Release Factors.
Cell Rep, 17, 2016
|
|
4M4P
 
 | | Crystal structure of EPHA4 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin type-A receptor 4 | | Authors: | Xu, K, Tsvetkova-Robev, D, Xu, Y, Goldgur, Y, Chan, Y.-P, Himanen, J.P, Nikolov, D.B. | | Deposit date: | 2013-08-07 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Insights into Eph receptor tyrosine kinase activation from crystal structures of the EphA4 ectodomain and its complex with ephrin-A5.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5D8B
 
 | |
8ABD
 
 | |
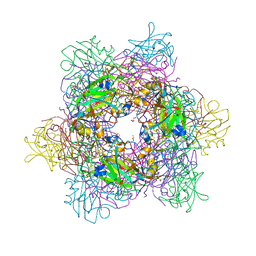
8ABN
 
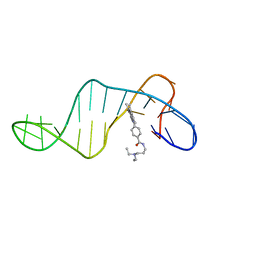 | |
5E81
 
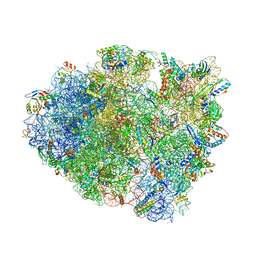 | | Structure of T. thermophilus 70S ribosome complex with mRNA and tRNALys in the A-site with wobble pair | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Khusainov, I, Yusupov, M, Yusupova, G. | | Deposit date: | 2015-10-13 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code.
Nat Commun, 7, 2016
|
|
3LXV
 
 | | Tyrosine 447 of Protocatechuate 3,4-Dioxygenase Controls Efficient Progress Through Catalysis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-NITROCATECHOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Purpero, V.M, Lipscomb, J.D. | | Deposit date: | 2010-02-25 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tyrosine 447 of Protocatechuate 3,4-Dioxygenase Controls Efficient Progress Through Catalysis
To be Published, 2010
|
|
5DOX
 
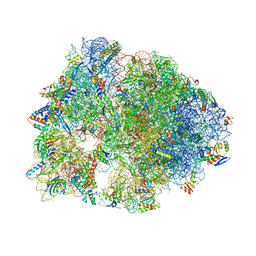 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Hygromycin-A at 3.1A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Starosta, A.L, Juette, M.F, Altman, R.B, Terry, D.S, Lu, W, Burnett, B.J, Dinos, G, Reynolds, K, Blanchard, S.C, Steitz, T.A, Wilson, D.N. | | Deposit date: | 2015-09-11 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A.
Mol.Cell, 58, 2015
|
|
8Z0T
 
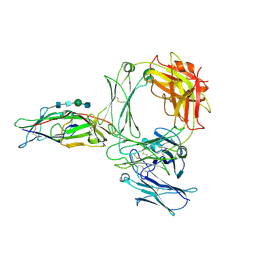 | | Structure of the human ige-fc bound to its high affinity receptor fc(epsilon) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Du, S, Deng, M.J, Xiao, J.Y. | | Deposit date: | 2024-04-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural insights into the high-affinity IgE receptor Fc epsilon RI complex.
Nature, 2024
|
|
8Y84
 
 | | Structure of the high affinity receptor fc(epsilon)ri TM | | Descriptor: | CHOLESTEROL HEMISUCCINATE, High affinity immunoglobulin epsilon receptor subunit alpha, High affinity immunoglobulin epsilon receptor subunit beta, ... | | Authors: | Du, S, Deng, M.J, Xiao, J.Y. | | Deposit date: | 2024-02-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural insights into the high-affinity IgE receptor Fc epsilon RI complex.
Nature, 2024
|
|
8ZGS
 
 | |
5DGE
 
 | | Coping with proline stalling: structural basis of hypusine-induced protein synthesis by the eukaryotic ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Melnikov, S, Mailliot, J, Shin, B.-S, Rigger, L, Yusupova, G, Micura, R, Dever, T.E, Yusupov, M. | | Deposit date: | 2015-08-27 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Coping with proline stalling: structural basis of hypusine-induced protein synthesis by the eukaryotic ribosome
To Be Published
|
|
5EL5
 
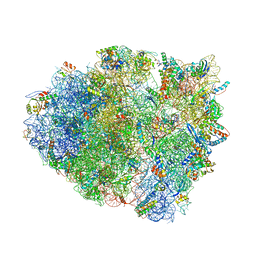 | | Structure of T. thermophilus 70S ribosome complex with mRNA and tRNALys in the A-site with a U-U mismatch in the second position | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Khusainov, I, Yusupov, M, Yusupova, G. | | Deposit date: | 2015-11-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code.
Nat Commun, 7, 2016
|
|
