2YBR
 
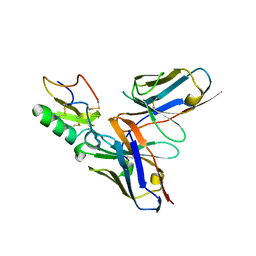 | | Crystal structure of the human derived single chain antibody fragment (scFv) 9004G in complex with Cn2 toxin from the scorpion Centruroides noxius Hoffmann | | Descriptor: | BETA-MAMMAL TOXIN CN2, SINGLE CHAIN ANTIBODY FRAGMENT 9004G | | Authors: | Canul-Tec, J.C, Riano-Umbarila, L, Rudino-Pinera, E, Becerril, B, Possani, L.D, Torres-Larios, A. | | Deposit date: | 2011-03-09 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis of Neutralization of the Major Toxic Component from the Scorpion Centruroides Noxius Hoffmann by a Human-Derived Single Chain Antibody Fragment
J.Biol.Chem., 286, 2011
|
|
2CDS
 
 | | LYSOZYME | | Descriptor: | PROTEIN (LYSOZYME (E.C.3.2.1.17)) | | Authors: | Kleywegt, G.J, Divne, C. | | Deposit date: | 1999-03-02 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Lysozyme
To be Published
|
|
2YBM
 
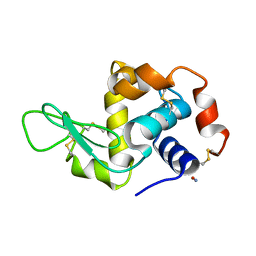 | |
3ZEZ
 
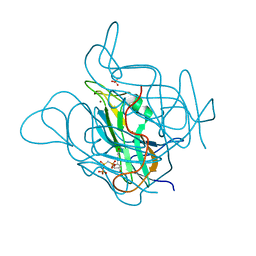 | | Phage dUTPases control transfer of virulence genes by a proto- oncogenic G protein-like mechanism.(Staphylococcus bacteriophage 80alpha dUTPase with dUPNHPP). | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPASE, MAGNESIUM ION, ... | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
2C7F
 
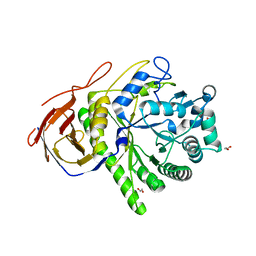 | | The Structure of a family 51 arabinofuranosidase, Araf51, from Clostridium thermocellum in complex with 1,5-alpha-L-Arabinotriose. | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-L-ARABINOFURANOSIDASE, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose, ... | | Authors: | Taylor, E.J, Smith, N.L, Turkenburg, J.P, D'Souza, S, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-11-23 | | Release date: | 2005-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insight Into the Ligand Specificity of a Thermostable Family 51 Arabinofuranosidase, Araf51, from Clostridium Thermocellum.
Biochem.J., 395, 2006
|
|
1YPF
 
 | | Crystal Structure of GuaC (BA5705) from Bacillus anthracis at 1.8 A Resolution | | Descriptor: | GMP reductase | | Authors: | Grenha, R, Levdikov, V.M, Blagova, E.V, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-31 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of GuaC (BA5705) from Bacillus anthracis at 1.8 A Resolution.
To be Published
|
|
5O1Z
 
 | |
2YID
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis alpha- ketoglutarate decarboxylase in complex with the enamine-ThDP intermediate | | Descriptor: | (4E)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-2(3H)-ylidene}-4-hydroxybutanoic acid, 2-OXOGLUTARATE DECARBOXYLASE, CALCIUM ION, ... | | Authors: | Wagner, T, Bellinzoni, M, Wehenkel, A.M, O'Hare, H.M, Alzari, P.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Functional Plasticity and Allosteric Regulation of Alpha-Ketoglutarate Decarboxylase in Central Mycobacterial Metabolism.
Chem.Biol., 18, 2011
|
|
4IU5
 
 | | Crystal structure of Leishmania mexicana arginase in complex with catalytic product L-ornithine | | Descriptor: | Arginase, GLYCEROL, L-ornithine, ... | | Authors: | D'Antonio, E.L, Ullman, B, Roberts, S.C, Gaur Dixit, U, Wilson, M.E, Hai, Y, Christianson, D.W. | | Deposit date: | 2013-01-19 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of arginase from Leishmania mexicana and implications for the inhibition of polyamine biosynthesis in parasitic infections.
Arch.Biochem.Biophys., 535, 2013
|
|
3ZHT
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD, first post-decarboxylation intermediate from 2-oxoadipate | | Descriptor: | (5S)-5-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-4-methyl-5-(2-{[(phosphonatooxy)phosphinato]oxy}ethyl)-1,3-thiazol-3-ium-2-yl}-5-hydroxypentanoate, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
1YS4
 
 | |
3ZO9
 
 | | The structure of Trehalose Synthase (TreS) of Mycobacterium smegmatis | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Caner, S, Nguyen, N, Aguda, A, Zhang, R, Pan, Y.T, Withers, S.G, Brayer, G.D. | | Deposit date: | 2013-02-21 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The Structure of the Mycobacterium Smegmatis Trehalose Synthase Reveals an Unusual Active Site Configuration and Acarbose-Binding Mode.
Glycobiology, 23, 2013
|
|
4IXT
 
 | | Structure of a 37-fold mutant of halohydrin dehalogenase (HheC) bound to ethyl (R)-4-cyano-3-hydroxybutyrate | | Descriptor: | CHLORIDE ION, Halohydrin dehalogenase, ethyl (3R)-4-cyano-3-hydroxybutanoate | | Authors: | Floor, R.J, Schallmey, M, Hauer, B, Breuer, M, Jekel, P.A, Wijma, H.J, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2013-01-28 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Biocatalytic and structural properties of a highly engineered halohydrin dehalogenase.
Chembiochem, 14, 2013
|
|
1YSD
 
 | | Yeast Cytosine Deaminase Double Mutant | | Descriptor: | CALCIUM ION, Cytosine deaminase, ZINC ION | | Authors: | Korkegian, A, Black, M.E, Baker, D, Stoddard, B.L. | | Deposit date: | 2005-02-08 | | Release date: | 2005-05-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational thermostabilization of an enzyme.
Science, 308, 2005
|
|
2CP4
 
 | |
1WB0
 
 | | specificity and affinity of natural product cyclopentapeptide inhibitor Argifin against human chitinase | | Descriptor: | ARGIFIN, CHITOTRIOSIDASE 1, GLYCEROL, ... | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Hodkinson, M, Adams, D.J, Shiomi, K, Omura, S, Van Aalten, D.M.F. | | Deposit date: | 2004-10-29 | | Release date: | 2005-01-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Specificity and Affinity of Natural Product Cyclopentapeptide Inhibitors Against Aspergillus Fumigatus, Human and Bacterial Chitinases
Chem.Biol., 12, 2005
|
|
4IQH
 
 | |
4EJY
 
 | | Structure of MBOgg1 in complex with high affinity DNA ligand | | Descriptor: | 3-Methyladenine DNA glycosylase, DNA (5'-D(*AP*GP*CP*GP*TP*CP*CP*AP*(3DR)P*GP*TP*CP*TP*AP*CP*C)-3'), DNA (5'-D(*T*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3'), ... | | Authors: | Jiang, T, Yu, H.J, Bi, L.J, Zhang, X.E, Yang, M.Z. | | Deposit date: | 2012-04-08 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of MBOgg1 in complex with two abasic DNA ligands
J.Struct.Biol., 181, 2013
|
|
3ZPI
 
 | | PikC D50N mutant in P21 space group | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME P450 HYDROXYLASE PIKC, FORMIC ACID, ... | | Authors: | Podust, L.M. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Recognition of Synthetic Substrates by P450 Pikc
To be Published
|
|
2YMP
 
 | | Chloroacetic acid complex bound L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | L-HALOACID DEHALOGENASE | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
4EF8
 
 | | Crystal structure of dihydroorotate dehydrogenase from Leishmania major in complex with Phenyl isothiocyanate | | Descriptor: | Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Pinheiro, M.P, Emery, F.S, Nonato, M.C. | | Deposit date: | 2012-03-29 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Target sites for the design of anti-trypanosomatid drugs based on the structure of dihydroorotate dehydrogenase.
Curr.Pharm.Des., 19, 2013
|
|
4IQI
 
 | | Crystal Structure of 7-cyano-7-deazaguanine Reductase, QueF from Vibrio cholerae O1 biovar El Tor complexed with cytosine | | Descriptor: | 6-AMINOPYRIMIDIN-2(1H)-ONE, CHLORIDE ION, NADPH-dependent 7-cyano-7-deazaguanine reductase, ... | | Authors: | Kim, Y, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-11 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of 7-cyano-7-deazaguanine Reductase, QueF from Vibrio cholerae O1 biovar El Tor complexed with cytosine
To be Published
|
|
4IQG
 
 | | Crystal structure of BPRO0239 oxidoreductase from Polaromonas sp. JS666 in NADP bound form | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, GLYCEROL, ... | | Authors: | Niedzialkowska, E, Majorek, K.A, Porebski, P.J, Al Obaidi, N, Hammonds, J, Hillerich, B, Seidel, R, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-01-11 | | Release date: | 2013-01-30 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of oxidoreductase from Polaromonas sp. in NADP bound form
To be Published
|
|
1NQT
 
 | | Crystal structure of bovine Glutamate dehydrogenase-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutamate dehydrogenase 1 | | Authors: | Banerjee, S, Schmidt, T, Fang, J, Stanley, C.A, Smith, T.J. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation
Biochemistry, 42, 2003
|
|
4EQV
 
 | |
