8QZJ
 
 | |
6NRG
 
 | | Crystal Structure of human PARP-1 ART domain bound to inhibitor UTT57 | | Descriptor: | 2-{[3-fluoro-4-(1H-tetrazol-5-yl)phenyl]methyl}-3-hydroxy-1-benzofuran-7-carboxamide, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Langelier, M.F, Pascal, J.M. | | Deposit date: | 2019-01-23 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Synthesis of Poly(ADP-ribose) Polymerase Inhibitors: Impact of Adenosine Pocket-Binding Motif Appendage to the 3-Oxo-2,3-dihydrobenzofuran-7-carboxamide on Potency and Selectivity.
J.Med.Chem., 62, 2019
|
|
8QZH
 
 | | Crystal structure of apo-PptT from Mycobacterium tuberculosis | | Descriptor: | 4'-phosphopantetheinyl transferase PptT, ACETATE ION, MAGNESIUM ION | | Authors: | Gavalda, S, Carivenc, C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2023-10-27 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic Cycle of Type II 4'-Phosphopantetheinyl Transferases
Acs Catalysis, 14, 2024
|
|
2XN2
 
 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with galactose | | Descriptor: | ALPHA-GALACTOSIDASE, GLYCEROL, IMIDAZOLE, ... | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
6NZM
 
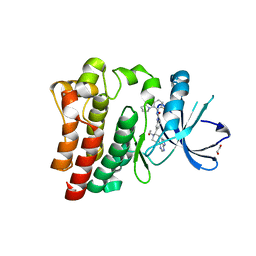 | | Brutons tyrosine kinase in complex with compound 50. | | Descriptor: | 1,2-ETHANEDIOL, N-[2-fluoro-6-(pyrrolidin-1-yl)phenyl]-N'-{3-[(2R)-1-(2-hydroxyethyl)-4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperazin-2-yl]phenyl}urea, Tyrosine-protein kinase BTK | | Authors: | Marcotte, D.J. | | Deposit date: | 2019-02-14 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Optimization of novel reversible Bruton's tyrosine kinase inhibitors identified using Tethering-fragment-based screens.
Bioorg.Med.Chem., 27, 2019
|
|
7FZ5
 
 | | Crystal Structure of human FABP4 in complex with rac-(2R)-1-[[3-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl]pyrrolidine-2-carboxylic acid | | Descriptor: | 1-{[(3M)-3-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl}-D-proline, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Neidhart, W, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
7FW9
 
 | | Crystal Structure of human FABP4 in complex with 2-[(3-ethoxycarbonyl-4,5,6,7-tetrahydro-1-benzothiophen-2-yl)carbamoyl]cyclopentene-1-carboxylic acid | | Descriptor: | 2-{[3-(ethoxycarbonyl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl}cyclopent-1-ene-1-carboxylic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Ceccarelli-Simona, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
1EXI
 
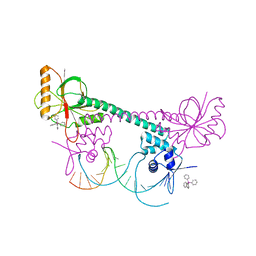 | | CRYSTAL STRUCTURE OF TRANSCRIPTION ACTIVATOR BMRR, FROM B. SUBTILIS, BOUND TO 21 BASE PAIR BMR OPERATOR AND TPSB | | Descriptor: | DNA (5'-D(*AP*CP*CP*CP*TP*CP*CP*CP*CP*TP*TP*AP*GP*GP*GP*GP*AP*GP*GP*GP*T)-3'), MULTIDRUG-EFFLUX TRANSPORTER REGULATOR, TETRAPHENYLANTIMONIUM ION, ... | | Authors: | Zheleznova-Heldwein, E.E, Brennan, R.G. | | Deposit date: | 2000-05-02 | | Release date: | 2001-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Crystal structure of the transcription activator BmrR bound to DNA and a drug.
Nature, 409, 2001
|
|
5KYJ
 
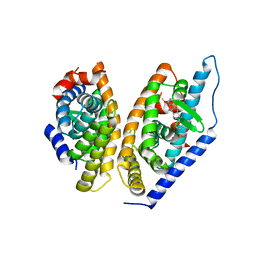 | | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core | | Descriptor: | (6~{R})-5-(5-fluoranyl-2-methoxy-pyrimidin-4-yl)-2-(3-methylsulfonylphenyl)-6-propan-2-yl-4,6-dihydropyrrolo[3,4-c]pyrazole, Oxysterols receptor LXR-beta, Retinoic acid receptor RXR-beta | | Authors: | Chen, G, McKeever, B.M. | | Deposit date: | 2016-07-21 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
7V0F
 
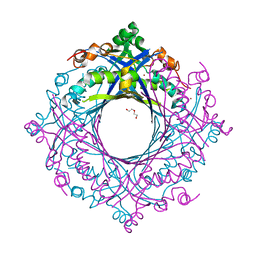 | | Structure of 6-carboxy-5,6,7,8-tetrahydropterin synthase paralog QueD2 from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, 6-carboxy-5,6,7,8-tetrahydropterin synthase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jordan, M.R, Gonzalez-Gutierrez, G, Giedroc, D.P. | | Deposit date: | 2022-05-10 | | Release date: | 2022-12-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Metal retention and replacement in QueD2 protect queuosine-tRNA biosynthesis in metal-starved Acinetobacter baumannii.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5L09
 
 | | Crystal Structure of Quorum-Sensing Transcriptional Activator from Yersinia enterocolitica in complex with 3-oxo-N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide | | Descriptor: | 1,2-ETHANEDIOL, 3-oxo-N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide, ACETIC ACID, ... | | Authors: | Kim, Y, Chhor, G, Jedrzejczak, R, Winans, S.C, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-26 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Quorum-Sensing Transcriptional Activator from Yersinia enterocolitica
To Be Published
|
|
5KYA
 
 | | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core | | Descriptor: | Oxysterols receptor LXR-beta, Retinoic acid receptor RXR-beta, [2-[(6~{R})-2-(3-methylsulfonylphenyl)-6-propan-2-yl-4,6-dihydropyrrolo[3,4-c]pyrazol-5-yl]-4-(trifluoromethyl)pyrimidin-5-yl]methanol | | Authors: | Chen, G, McKeever, B.M. | | Deposit date: | 2016-07-21 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
6ALB
 
 | | CREBBP bromodomain in complex with Cpd 30 (1-(3-(3-(1-methyl-1H-pyrazol-4-yl)isoquinolin-8-yl)-1-(tetrahydro-2H-pyran-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | Descriptor: | 1-{3-[3-(1-methyl-1H-pyrazol-4-yl)isoquinolin-8-yl]-1-(oxan-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CREB-binding protein, ... | | Authors: | Murray, J.M. | | Deposit date: | 2017-08-07 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Design and Synthesis of A Biaryl Series As Inhibitors for the Bromodomains of CBP/P300
To be published
|
|
8AX1
 
 | |
2XN1
 
 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-GALACTOSIDASE, GLYCEROL | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
8UFR
 
 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 4-methyl-7-(4-methyl-2,3,4,5-tetrahydrobenzo[f][1,4]oxazepin-7-yl)quinolin-2-amine dihydrochloride | | Descriptor: | (7M)-4-methyl-7-(4-methyl-2,3,4,5-tetrahydro-1,4-benzoxazepin-7-yl)quinolin-2-amine, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2023-10-04 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.871 Å) | | Cite: | Crystallographic and Computational Insights into Isoform-Selective Dynamics in Nitric Oxide Synthase.
Biochemistry, 63, 2024
|
|
5LPS
 
 | | tRNA guanine Transglycosylase (TGT) in co-crystallized complex (space group C2) with 6-amino-2-(methylamino)-4-(2-((2R,3R,4S,5R,6S)-3,4,5,6-tetramethoxytetrahydro-2H-pyran-2-yl)ethyl)-1H-imidazo[4,5-g]quinazolin-8(7H)-one | | Descriptor: | 1,2-ETHANEDIOL, 6-azanyl-2-(methylamino)-4-[2-[(2~{R},3~{R},4~{S},5~{R},6~{S})-3,4,5,6-tetramethoxyoxan-2-yl]ethyl]-1,7-dihydroimidazo[4,5-g]quinazolin-8-one, Queuine tRNA-ribosyltransferase, ... | | Authors: | Ehrmann, F.R, Heine, A, Klebe, G. | | Deposit date: | 2016-08-14 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Carbohydrate-based Inhibitors targeting the Ribose-34 pocket of Z.mobilis TGT and changing the oligomeric state of the homodimer
To be published
|
|
4TWS
 
 | | Gadolinium Derivative of Tetragonal Hen Egg-White Lysozyme at 1.45 A Resolution | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Holton, J.M, Classen, S, Frankel, K.A, Tainer, J.A. | | Deposit date: | 2014-07-01 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The R-factor gap in macromolecular crystallography: an untapped potential for insights on accurate structures.
Febs J., 281, 2014
|
|
5MSB
 
 | | Crystal structure of human carbonic anhydrase isozyme XII with 2,3,5,6-tetrafluoro-4[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 2,3,5,6-tetrafluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2017-01-02 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure correlations with the intrinsic thermodynamics of human carbonic anhydrase inhibitor binding.
PeerJ, 6, 2018
|
|
5LJ2
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH 5-(5-aminopyridin-3-yl)-8-(((3R,4R)-3-((1,1-dioxidotetrahydro-2H-thiopyran-4-yl)methoxy)piperidin-4-yl)amino)-3-methyl-1,7-naphthyridin-2(1H)-one | | Descriptor: | 1,2-ETHANEDIOL, 5-(5-aminopyridin-3-yl)-8-(((3R,4R)-3-((1,1-dioxidotetrahydro-2H-thiopyran-4-yl)methoxy)piperidin-4-yl)amino)-3-methyl-1,7-naphthyridin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Chung, C, Bamborough, P, Demont, E. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | A Chemical Probe for the ATAD2 Bromodomain.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
1AXV
 
 | | SOLUTION NMR STRUCTURE OF THE [BP]DA ADDUCT OPPOSITE DT IN A DNA DUPLEX, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(CTCTC-[BP]A-CTTCC)D(GGAAGTGAGAG) | | Authors: | Mao, B, Gu, Z, Gorin, A.A, Chen, J, Hingerty, B.E, Amid, S, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 1997-10-21 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the (+)-cis-anti-benzo[a]pyrene-dA ([BP]dA) adduct opposite dT in a DNA duplex.
Biochemistry, 38, 1999
|
|
1AXR
 
 | | COOPERATIVITY BETWEEN HYDROGEN-BONDING AND CHARGE-DIPOLE INTERACTIONS IN THE INHIBITION OF BETA-GLYCOSIDASES BY AZOLOPYRIDINES: EVIDENCE FROM A STUDY WITH GLYCOGEN PHOSPHORYLASE B | | Descriptor: | 4,5,6-TRIHYDROXY-7-HYDROXYMETHYL-4,5,6,7-TETRAHYDRO-1H-[1,2,3]TRIAZOLO[1,5-A]PYRIDIN-8-YLIUM, GLYCOGEN PHOSPHORYLASE, PHOSPHATE ION, ... | | Authors: | Oikonomakos, N.G, Heightman, T.D. | | Deposit date: | 1997-10-20 | | Release date: | 1998-01-28 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cooperative interactions of the catalytic nucleophile and the catalytic acid in the inhibition of beta-glycosidases. Calculations and their validation by comparative kinetic and structural studies of the inhibition of glycogen phosphorylase b.
HELV.CHIM.ACTA, 81, 1998
|
|
5LPT
 
 | | tRNA guanine Transglycosylase (TGT) in co-crystallized complex (space group P21) with 6-amino-2-(methylamino)-4-(2-((2R,3R,4S,5R,6S)-3,4,5,6-tetramethoxytetrahydro-2H-pyran-2-yl)ethyl)-1H-imidazo[4,5-g]quinazolin-8(7H)-one | | Descriptor: | 6-azanyl-2-(methylamino)-4-[2-[(2~{R},3~{R},4~{S},5~{R},6~{S})-3,4,5,6-tetramethoxyoxan-2-yl]ethyl]-1,7-dihydroimidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Ehrmann, F.R, Heine, A, Klebe, G. | | Deposit date: | 2016-08-14 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Carbohydrate-based Inhibitors targeting the Ribose-34 pocket of Z.mobilis TGT and changing the oligomeric state of the homodimer
To be published
|
|
5NHS
 
 | | The crystal structure of Xanthomonas albilineans N5, N10-methylenetetrahydrofolate dehydrogenase-cyclohydrolase (FolD) | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional protein FolD | | Authors: | Bueno, R.V, Guido, R.V.C, Maluf, F.V, Lima, G.M.A, Oliveira, A.A. | | Deposit date: | 2017-03-22 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The crystal structure of Xanthomonas albilineans N5, N10-methylenetetrahydrofolate dehydrogenase-cyclohydrolase (FolD)
To Be Published
|
|
5J91
 
 | | Structure of the Wild-type 70S E coli ribosome bound to Tigecycline | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-04-08 | | Release date: | 2016-07-06 | | Last modified: | 2016-08-03 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
