2RB8
 
 | |
3VBT
 
 | |
6LDW
 
 | |
3VC4
 
 | |
6OV9
 
 | |
6OVS
 
 | | Coiled-coil Trimer with Glu:Tyr:Lys Triad | | Descriptor: | Coiled-coil Trimer with Glu:Tyr:Lys Triad, PENTAETHYLENE GLYCOL | | Authors: | Smith, M.S, Stern, K.L, Billings, W.M, Price, J.L. | | Deposit date: | 2019-05-08 | | Release date: | 2020-04-29 | | Last modified: | 2020-05-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Context-Dependent Stabilizing Interactions among Solvent-Exposed Residues along the Surface of a Trimeric Helix Bundle.
Biochemistry, 59, 2020
|
|
6LDV
 
 | |
6OSD
 
 | |
6OLO
 
 | |
2RLZ
 
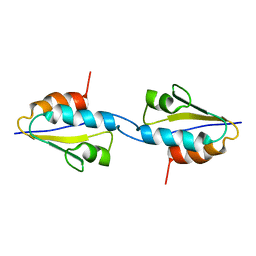 | | Solid-State MAS NMR structure of the dimer Crh | | Descriptor: | HPr-like protein crh | | Authors: | Loquet, A, Bardiaux, B, Gardiennet, C, Blanchet, C, Baldus, M, Nilges, M, Malliavin, T, Bockmann, A. | | Deposit date: | 2007-09-04 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-29 | | Method: | SOLID-STATE NMR | | Cite: | 3D structure determination of the Crh protein from highly ambiguous solid-state NMR restraints.
J.Am.Chem.Soc., 130, 2008
|
|
3VBV
 
 | |
6OVV
 
 | |
3VBQ
 
 | |
7YTO
 
 | |
3VBY
 
 | |
5I8B
 
 | | CBP in complex with Cpd23 ((R)-6-(3-(benzyloxy)phenyl)-4-methyl-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one) | | Descriptor: | (4R)-6-[3-(benzyloxy)phenyl]-4-methyl-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5218 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
6LRB
 
 | |
6LT0
 
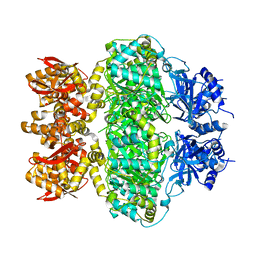
 | | cryo-EM structure of C9ORF72-SMCR8-WDR41 | | Descriptor: | Guanine nucleotide exchange C9orf72, Guanine nucleotide exchange protein SMCR8, WD repeat-containing protein 41 | | Authors: | Tang, D, Sheng, J, Xu, L, Zhan, X, Yan, C, Qi, S. | | Deposit date: | 2020-01-21 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of C9ORF72-SMCR8-WDR41 reveals the role as a GAP for Rab8a and Rab11a.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7Q3K
 
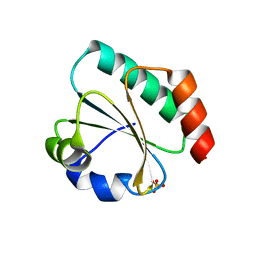 | | Computationally designed thioredoxin subjected to stability optimizing mutations. | | Descriptor: | SULFATE ION, eMM9 | | Authors: | Norrild, R.K, Johansson, K.E, O'Shea, C, Lindorff-Larsen, K, Winther, J.R, Morth, J.P. | | Deposit date: | 2021-10-27 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Increasing protein stability by inferring substitution effects from high-throughput experiments.
Cell Rep Methods, 2, 2022
|
|
4V7Q
 
 | | Atomic model of an infectious rotavirus particle | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Core scaffold protein, ... | | Authors: | Settembre, E.C, Chen, J.Z, Dormitzer, P.R, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2010-05-13 | | Release date: | 2014-07-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic model of an infectious rotavirus particle.
Embo J., 30, 2011
|
|
6M2V
 
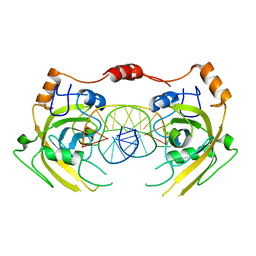 | | Crystal structure of UHRF1 SRA complexed with fully-mCHG DNA. | | Descriptor: | DNA (5'-D(*TP*CP*AP*CP*GP*(5CM)P*TP*GP*CP*GP*TP*GP*A)-3'), E3 ubiquitin-protein ligase UHRF1 | | Authors: | Abhishek, S, Nakarakanti, N.K, Deeksha, W, Rajakumara, E. | | Deposit date: | 2020-03-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanistic insights into recognition of symmetric methylated cytosines in CpG and non-CpG DNA by UHRF1 SRA.
Int.J.Biol.Macromol., 170, 2021
|
|
6PA7
 
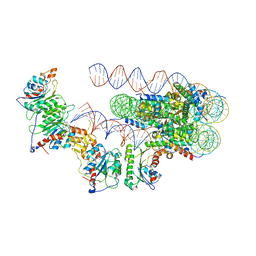 | | The cryo-EM structure of the human DNMT3A2-DNMT3B3 complex bound to nucleosome. | | Descriptor: | CHLORIDE ION, DNA (167-MER), DNA (cytosine-5)-methyltransferase 3A, ... | | Authors: | Xu, T.H, Liu, M, Zhou, X.E, Liang, G, Zhao, G, Xu, H.E, Melcher, K, Jones, P.A. | | Deposit date: | 2019-06-11 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structure of nucleosome-bound DNA methyltransferases DNMT3A and DNMT3B.
Nature, 586, 2020
|
|
6G65
 
 | |
6G6C
 
 | |
6FP9
 
 | | Crystal structure of anti-mTFP1 DARPin 1238_E11 | | Descriptor: | 1,2-ETHANEDIOL, DARPin 1238_E11, SULFATE ION | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
