3RU4
 
 | | Crystal structure of the Bowman-Birk serine protease inhibitor BTCI in complex with trypsin and chymotrypsin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, Bowman-Birk type seed trypsin and chymotrypsin inhibitor, ... | | Authors: | Esteves, G.F, Santos, C.R, Ventura, M.M, Barbosa, J.A.R.G, Freitas, S.M. | | Deposit date: | 2011-05-04 | | Release date: | 2012-08-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of the Bowman-Birk serine protease inhibitor BTCI in complex with trypsin and chymotrypsin
To be Published
|
|
1OAD
 
 | | Glucose isomerase from Streptomyces rubiginosus in P21212 crystal form | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Ramagopal, U.A, Dauter, M, Dauter, Z. | | Deposit date: | 2003-01-08 | | Release date: | 2003-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sad Manganese in Two Crystal Forms of Glucose Isomerase
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3WL2
 
 | | Monoclinic Lysozyme at 0.96 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Lysozyme C, NITRATE ION, ... | | Authors: | Matsumoto, T, Yamano, A, Hasegawa, T, Maeyama, M. | | Deposit date: | 2013-11-06 | | Release date: | 2014-11-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Evaluation of Rigaku XtaLAB P200
To be Published
|
|
3S3Q
 
 | | Structure of cathepsin B1 from Schistosoma mansoni in complex with K11017 inhibitor | | Descriptor: | ACETATE ION, Cathepsin B-like peptidase (C01 family), N~2~-(morpholin-4-ylcarbonyl)-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-leucinamide | | Authors: | Rezacova, P, Jilkova, A, Brynda, J, Horn, M, Mares, M. | | Deposit date: | 2011-05-18 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Inhibition of Cathepsin B Drug Target from the Human Blood Fluke, Schistosoma mansoni.
J.Biol.Chem., 286, 2011
|
|
1QJX
 
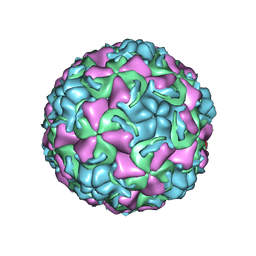 | | HUMAN RHINOVIRUS 16 COAT PROTEIN IN COMPLEX WITH ANTIVIRAL COMPOUND WIN68934 | | Descriptor: | 2,6-DIMETHYL-1-(3-[3-METHYL-5-ISOXAZOLYL]-PROPANYL)-4-[4-METHYL-2H-TETRAZOL-2-YL]-PHENOL, MYRISTIC ACID, PROTEIN VP1, ... | | Authors: | Hadfield, A.T, Diana, G.D, Rossmann, M.G. | | Deposit date: | 1999-07-06 | | Release date: | 1999-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of Three Structurally Related Antiviral Compounds in Complex with Human Rhinovirus 16
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2ASF
 
 | | Crystal structure of the conserved hypothetical protein Rv2074 from Mycobacterium tuberculosis 1.6 A | | Descriptor: | CITRIC ACID, Hypothetical protein Rv2074, SODIUM ION | | Authors: | Biswal, B.K, Au, K, Cherney, M.M, Garen, C, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The molecular structure of Rv2074, a probable pyridoxine 5'-phosphate oxidase from Mycobacterium tuberculosis, at 1.6 angstroms resolution.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
3N1W
 
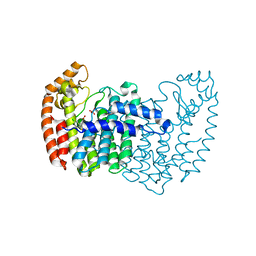 | | Human FPPS COMPLEX WITH FBS_02 | | Descriptor: | (5-chloro-1-benzothiophen-3-yl)acetic acid, FARNESYL PYROPHOSPHATE SYNTHASE, PHOSPHATE ION | | Authors: | Rondeau, J.-M. | | Deposit date: | 2010-05-17 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Allosteric non-bisphosphonate FPPS inhibitors identified by fragment-based discovery.
Nat.Chem.Biol., 6, 2010
|
|
3CNH
 
 | |
2WDB
 
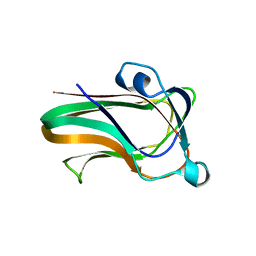 | | A family 32 carbohydrate-binding module, from the Mu toxin produced by Clostridium perfringens, in complex with beta-D-glcNAc-beta(1,2) mannose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, CALCIUM ION, HYALURONOGLUCOSAMINIDASE | | Authors: | Ficko-Blean, E, Boraston, A.B. | | Deposit date: | 2009-03-23 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | N-Acetylglucosamine Recognition by a Family 32 Carbohydrate-Binding Module from Clostridium Perfringens Nagh.
J.Mol.Biol., 390, 2009
|
|
1HFF
 
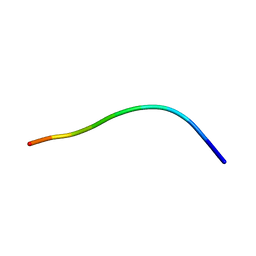 | | NMR solution structures of the vMIP-II 1-10 peptide from Kaposi's sarcoma-associated herpesvirus. | | Descriptor: | VIRAL MACROPHAGE INFLAMMATORY PROTEIN-II | | Authors: | Crump, M.P, Elisseeva, E, Gong, J.H, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 2000-12-01 | | Release date: | 2000-12-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure/Function of Human Herpesvirus-8 Mip-II (1-71) and the Antagonist N-Terminal Segment (1-10)
FEBS Lett., 489, 2001
|
|
1HFG
 
 | | NMR solution structure of vMIP-II 1-71 from Kaposi's sarcoma-associated herpesvirus (minimized average structure). | | Descriptor: | VIRAL MACROPHAGE INFLAMMATORY PROTEIN-II | | Authors: | Crump, M.P, Elisseeva, E, Gong, J.-H, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 2000-12-01 | | Release date: | 2001-01-07 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structure/Function of Human Herpesvirus-8 Mip-II (1-71) and the Antagonist N-Terminal Segment (1-10)
FEBS Lett., 489, 2001
|
|
3CR4
 
 | | X-ray structure of bovine Pnt,Ca(2+)-S100B | | Descriptor: | 1,5-BIS(4-AMIDINOPHENOXY)PENTANE, CALCIUM ION, Protein S100-B | | Authors: | Charpentier, T.H. | | Deposit date: | 2008-04-04 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Divalent metal ion complexes of S100B in the absence and presence of pentamidine.
J.Mol.Biol., 382, 2008
|
|
2QEU
 
 | |
3UV5
 
 | | Crystal Structure of the tandem bromodomains of human Transcription initiation factor TFIID subunit 1 (TAF1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Transcription initiation factor TFIID subunit 1 | | Authors: | Filippakopoulos, P, Felletar, I, Picaud, S, Keates, T, Muniz, J, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-29 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3V4Y
 
 | | Crystal Structure of the first Nuclear PP1 holoenzyme | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Nuclear inhibitor of protein phosphatase 1, ... | | Authors: | Page, R, Peti, W, O'Connell, N.E, Nichols, S. | | Deposit date: | 2011-12-15 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | The Molecular Basis for Substrate Specificity of the Nuclear NIPP1:PP1 Holoenzyme.
Structure, 20, 2012
|
|
7JJG
 
 | |
2QNP
 
 | | HIV-1 Protease in complex with a iodo decorated pyrrolidine-based inhibitor | | Descriptor: | CHLORIDE ION, Gag-Pol polyprotein (Pr160Gag-Pol), N,N'-(3S,4S)-pyrrolidine-3,4-diylbis[N-(4-iodobenzyl)benzenesulfonamide] | | Authors: | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2007-07-19 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structure-Guided Design of C2-Symmetric HIV-1 Protease Inhibitors Based on a Pyrrolidine Scaffold.
J.Med.Chem., 51, 2008
|
|
4HKL
 
 | | Crystal Structures of Mutant Endo-beta-1,4-xylanase II Complexed with substrate (1.15 A) and Products (1.6 A) | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Langan, P, Wan, Q, Coates, L, Kovalevsky, A. | | Deposit date: | 2012-10-15 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1GOO
 
 | | Thermostable xylanase I from Thermoascus aurantiacus - Cryocooled glycerol complex | | Descriptor: | ENDO-1,4-BETA-XYLANASE, GLYCEROL | | Authors: | Eckert, K, Andrei, C, Larsen, S, Lo Leggio, L. | | Deposit date: | 2001-10-22 | | Release date: | 2001-12-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Substrate Specificity and Subsite Mobility in T. Aurantiacus Xylanase 10A
FEBS Lett., 509, 2001
|
|
3N1V
 
 | | Human FPPS COMPLEX WITH FBS_01 | | Descriptor: | (5-chloro-3-methyl-1-benzothiophen-2-yl)acetic acid, FARNESYL PYROPHOSPHATE SYNTHASE, PHOSPHATE ION | | Authors: | Rondeau, J.-M. | | Deposit date: | 2010-05-17 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Allosteric non-bisphosphonate FPPS inhibitors identified by fragment-based discovery.
Nat.Chem.Biol., 6, 2010
|
|
1LPP
 
 | |
1H95
 
 | | Solution structure of the single-stranded DNA-binding Cold Shock Domain (CSD) of human Y-box protein 1 (YB1) determined by NMR (10 lowest energy structures) | | Descriptor: | Y-BOX BINDING PROTEIN | | Authors: | Kloks, C.P.A.M, Spronk, C.A.E.M, Hoffmann, A, Vuister, G.W, Grzesiek, S, Hilbers, C.W. | | Deposit date: | 2001-02-23 | | Release date: | 2002-02-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure and DNA-Binding Properties of the Cold-Shock Domain of the Human Y-Box Protein Yb-1.
J.Mol.Biol., 316, 2002
|
|
2CFZ
 
 | | Crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa, in complex with 1-dodecanol | | Descriptor: | 1-DODECANOL, DI(HYDROXYETHYL)ETHER, SDS HYDROLASE SDSA1, ... | | Authors: | Hagelueken, G, Adams, T.M, Wiehlmann, L, Widow, U, Kolmar, H, Tuemmler, B, Heinz, D.W, Schubert, W.-D. | | Deposit date: | 2006-02-26 | | Release date: | 2006-04-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Crystal Structure of Sdsa1, an Alkylsulfatase from Pseudomonas Aeruginosa, Defines a Third Class of Sulfatases.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
7JSP
 
 | | Crystal structure of the second bromodomain (BD2) of human TAF1 bound to ATR kinase inhibitor AZD6738 | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-{1-[(R)-amino(hydroxy)methyl-lambda~4~-sulfanyl]cyclopropyl}-6-[(3R)-3-methylmorpholin-4-yl]pyrimidin-2-yl)-1H-pyrrolo[2,3-b]pyridine, SULFATE ION, ... | | Authors: | Karim, M.R, Schonbrunn, E. | | Deposit date: | 2020-08-15 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Dual TAF1-ATR Inhibitors and Ligand-Induced Structural Changes of the TAF1 Tandem Bromodomain.
J.Med.Chem., 65, 2022
|
|
7JKY
 
 | | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with YF3-126 | | Descriptor: | Bromodomain-containing protein 4, N-(1-[1,1-di(pyridin-2-yl)ethyl]-6-{1-methyl-6-oxo-5-[(piperidin-4-yl)amino]-1,6-dihydropyridin-3-yl}-1H-indol-4-yl)ethanesulfonamide, SODIUM ION | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | Deposit date: | 2020-07-29 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Novel Pyrrolopyridone Bromodomain and Extra-Terminal Motif (BET) Inhibitors Effective in Endocrine-Resistant ER+ Breast Cancer with Acquired Resistance to Fulvestrant and Palbociclib.
J.Med.Chem., 63, 2020
|
|
