8ZC2
 
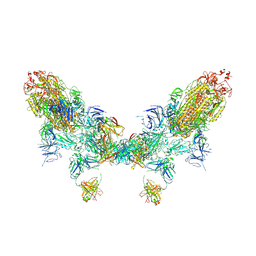 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with D1F6 Fab, head-to-head aggregate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (7.82 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
4P65
 
 | | Crystal structure of an cyclohexylalanine substituted insulin analog. | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Pandyarajan, V, Wan, Z, Weiss, M.A. | | Deposit date: | 2014-03-21 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Aromatic Anchor at an Invariant Hormone-Receptor Interface: FUNCTION OF INSULIN RESIDUE B24 WITH APPLICATION TO PROTEIN DESIGN.
J.Biol.Chem., 289, 2014
|
|
6LD4
 
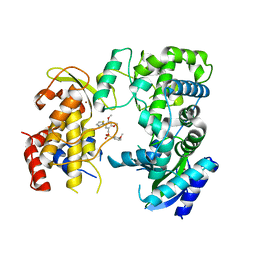 | | Zika NS5 polymerase domain | | Descriptor: | 2,4-dimethoxy-5-thiophen-2-yl-benzoic acid, 3-methoxybenzenesulfonamide, RNA-directed RNA polymerase NS5, ... | | Authors: | El Sahili, A, Lescar, J. | | Deposit date: | 2019-11-20 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Non-nucleoside Inhibitors of Zika Virus RNA-Dependent RNA Polymerase.
J.Virol., 94, 2020
|
|
6LD3
 
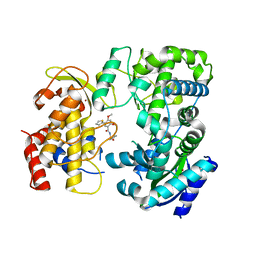 | | Zika NS5 polymerase domain | | Descriptor: | 2,4-dimethoxy-5-thiophen-2-yl-benzoic acid, RNA-directed RNA polymerase NS5, ZINC ION, ... | | Authors: | El Sahili, A, Lescar, J. | | Deposit date: | 2019-11-20 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Non-nucleoside Inhibitors of Zika Virus RNA-Dependent RNA Polymerase.
J.Virol., 94, 2020
|
|
4PIC
 
 | |
6L2Q
 
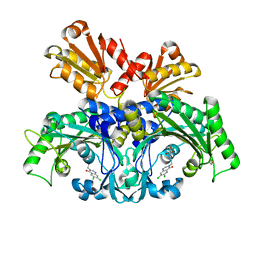 | | Threonyl-tRNA synthetase from Salmonella enterica in complex with an inhibitor | | Descriptor: | (2S,3R)-2-azanyl-N-[(E)-4-[6,7-bis(chloranyl)-4-oxidanylidene-quinazolin-3-yl]but-2-enyl]-3-oxidanyl-butanamide, 1,2-ETHANEDIOL, Threonine--tRNA ligase, ... | | Authors: | Guo, J, Chen, B, Zhou, H. | | Deposit date: | 2019-10-05 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of novel tRNA-amino acid dual-site inhibitors against threonyl-tRNA synthetase by fragment-based target hopping.
Eur.J.Med.Chem., 187, 2019
|
|
6LJK
 
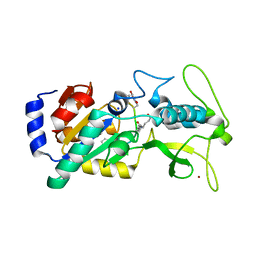 | |
6LIS
 
 | | ASFV dUTPase in complex with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, E165R | | Authors: | Liang, R, Peng, G.Q. | | Deposit date: | 2019-12-12 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development.
J.Biol.Chem., 296, 2020
|
|
6LJI
 
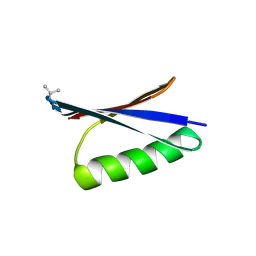 | | X-ray structure of synthetic GB1 domain with mutations K10(DVA), T11V | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Penmatsa, A, Chatterjee, J, Majumder, P, Khatri, B. | | Deposit date: | 2019-12-16 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.843 Å) | | Cite: | Increasing protein stability by engineering the n -> pi * interaction at the beta-turn.
Chem Sci, 11, 2020
|
|
6LJ3
 
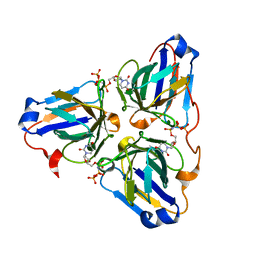 | |
6LJO
 
 | | African swine fever virus dUTPase | | Descriptor: | E165R | | Authors: | Liang, R, Peng, G.Q. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development.
J.Biol.Chem., 296, 2020
|
|
6L0L
 
 | | Hydra-1ubq de nova designed by Hydra based on ubiquitin | | Descriptor: | Hydra-1ubq | | Authors: | Ouyang, B. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multiobjective heuristic algorithm for de novo protein design in a quantified continuous sequence space.
Comput Struct Biotechnol J, 19, 2021
|
|
6M08
 
 | | Crystal structure of Lp-PLA2 in complex with a novel covalent inhibitor. | | Descriptor: | (2S)-2-[(Z)-3-[2-(diethylamino)ethyl-[[4-[4-(trifluoromethyl)phenyl]phenyl]methyl]amino]-1-oxidanyl-3-oxidanylidene-prop-1-enyl]pyrrolidine-1-carboxylic acid, Platelet-activating factor acetylhydrolase | | Authors: | Hu, H.C, Xu, Y.C. | | Deposit date: | 2020-02-20 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identification of Highly Selective Lipoprotein-Associated Phospholipase A2 (Lp-PLA2) Inhibitors by a Covalent Fragment-Based Approach.
J.Med.Chem., 63, 2020
|
|
4PQ7
 
 | |
6LZ4
 
 | |
6LR7
 
 | |
6V1V
 
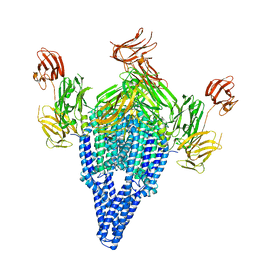 | | VIP3B (VIP3B_2160) adapted for crystallization | | Descriptor: | Vegetative insecticidal protein | | Authors: | Evdokimov, A.G, Zheng, M, Moshiri, F, Haas, J, Lowder, C. | | Deposit date: | 2019-11-21 | | Release date: | 2019-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.189 Å) | | Cite: | Crystal structure of a Vip3B family insecticidal protein reveals a new fold and a unique tetrameric assembly.
Protein Sci., 29, 2020
|
|
6VJ3
 
 | | Carbonic Anhydrase II in complex with pyrimidine-based inhibitor | | Descriptor: | 2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-N-(4-sulfamoylphenyl)acetamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Lomelino, C.L, McKenna, R. | | Deposit date: | 2020-01-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Inclusion of a 5-fluorouracil moiety in nitrogenous bases derivatives as human carbonic anhydrase IX and XII inhibitors produced a targeted action against MDA-MB-231 and T47D breast cancer cells.
Eur.J.Med.Chem., 190, 2020
|
|
6X0Q
 
 | |
6X0R
 
 | |
1EJ8
 
 | | CRYSTAL STRUCTURE OF DOMAIN 2 OF THE YEAST COPPER CHAPERONE FOR SUPEROXIDE DISMUTASE (LYS7) AT 1.55 A RESOLUTION | | Descriptor: | CALCIUM ION, LYS7 | | Authors: | Hall, L.T, Sanchez, R.J, Holloway, S.P, Zhu, H, Stine, J.E, Lyons, T.J, Demeler, B, Schirf, V, Hansen, J.C, Nersissian, A.M, Valentine, J.S, Hart, P.J. | | Deposit date: | 2000-03-01 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystallographic and analytical ultracentrifugation analyses of truncated and full-length yeast copper chaperones for SOD (LYS7): a dimer-dimer model of LYS7-SOD association and copper delivery.
Biochemistry, 39, 2000
|
|
3VBP
 
 | |
5KZQ
 
 | | Metabotropic Glutamate Receptor in complex with antagonist (1~{S},2~{R},3~{S},4~{S},5~{R},6~{R})-2-azanyl-3-[[3,4-bis(fluoranyl)phenyl]sulfanylmethyl]-4-oxidanyl-bicyclo[3.1.0]hexane-2,6-dicarboxylic acid | | Descriptor: | (1~{S},2~{R},3~{S},4~{S},5~{R},6~{R})-2-azanyl-3-[[3,4-bis(fluoranyl)phenyl]sulfanylmethyl]-4-oxidanyl-bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2 | | Authors: | Chappell, M.D, Li, R, Smith, S.C, Dressman, B.A, Tromiczak, E.G, Tripp, A.E, Blanco, M.-J, Vetman, T, Quimby, S.J, Matt, J, Britton, T, Fivush, A.M, Schkeryantz, J.M, Mayhugh, D, Erickson, J.A, Bures, M, Jaramillo, C, Carpintero, M, de Diego, J.E, Barberis, M, Garcia-Cerrada, S, Soriano, J.F, Antonysamy, S, Atwell, S, MacEwan, I, Condon, B, Bradley, C, Wang, J, Zhang, A, Conners, K, Groshong, C, Wasserman, S.R, Koss, J.W, Witkin, J.M, Li, X, Overshiner, C, Wafford, K.A, Seidel, W, Wang, X.-S, Heinz, B.A, Swanson, S, Catlow, J, Bedwell, D, Monn, J.A, Mitch, C.H, Ornstein, P. | | Deposit date: | 2016-07-25 | | Release date: | 2016-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of (1S,2R,3S,4S,5R,6R)-2-Amino-3-[(3,4-difluorophenyl)sulfanylmethyl]-4-hydroxy-bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid Hydrochloride (LY3020371HCl): A Potent, Metabotropic Glutamate 2/3 Receptor Antagonist with Antidepressant-Like Activity.
J. Med. Chem., 59, 2016
|
|
3V9H
 
 | |
5L1X
 
 | | Structure of the Human Metapneumovirus Fusion Protein in the Postfusion Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mas, V, Melero, J.A, McLellan, J.S. | | Deposit date: | 2016-07-29 | | Release date: | 2016-08-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Engineering, Structure and Immunogenicity of the Human Metapneumovirus F Protein in the Postfusion Conformation.
Plos Pathog., 12, 2016
|
|
