1DMS
 
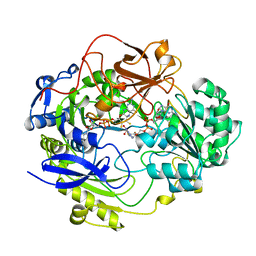 | | STRUCTURE OF DMSO REDUCTASE | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DMSO REDUCTASE, MOLYBDENUM (IV)OXIDE | | Authors: | Schneider, F, Loewe, J, Huber, R, Schindelin, H, Kisker, C, Knaeblein, J. | | Deposit date: | 1996-09-03 | | Release date: | 1998-07-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of dimethyl sulfoxide reductase from Rhodobacter capsulatus at 1.88 A resolution.
J.Mol.Biol., 263, 1996
|
|
1DPB
 
 | |
1DO2
 
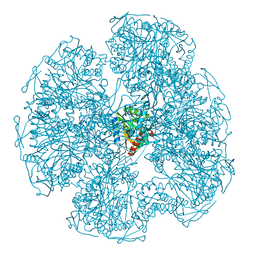
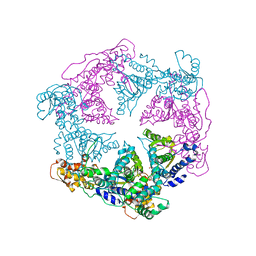 | | TRIGONAL CRYSTAL FORM OF HEAT SHOCK LOCUS U (HSLU) FROM ESCHERICHIA COLI | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PROTEIN (HEAT SHOCK LOCUS U) | | Authors: | Bochtler, M, Hartmann, C, Song, H.K, Bourenkov, G.P, Bartunik, H.D. | | Deposit date: | 1999-12-18 | | Release date: | 2000-02-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The structures of HsIU and the ATP-dependent protease HsIU-HsIV.
Nature, 403, 2000
|
|
1DDT
 
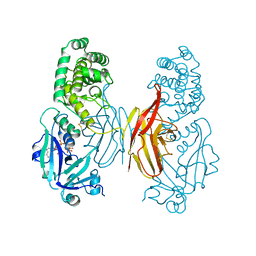 | |
1DIF
 
 | | HIV-1 PROTEASE IN COMPLEX WITH A DIFLUOROKETONE CONTAINING INHIBITOR A79285 | | Descriptor: | BETA-MERCAPTOETHANOL, HIV-1 PROTEASE, N-{1-BENZYL-2,2-DIFLUORO-3,3-DIHYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE | | Authors: | Silva, A.M, Cachau, R.E, Sham, H.L, Erickson, J.W. | | Deposit date: | 1995-10-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition and catalytic mechanism of HIV-1 aspartic protease.
J.Mol.Biol., 255, 1996
|
|
7CE9
 
 | | PQQ-soaked Apo-methanol dehydrogenase (MDH) from Methylococcus capsulatus (Bath) | | Descriptor: | CALCIUM ION, Methanol dehydrogenase [cytochrome c] subunit 2, Methanol dehydrogenase protein, ... | | Authors: | Chuankhayan, P, Chan, S.I, Nareddy, P.K.R, Tsai, I.K, Tsai, Y.F, Chen, K.H.-C, Yu, S.S.-F, Chen, C.J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Pyrroloquinoline Quinone-Dependent Hydride Transfer Chemistry from Spectroscopic and High-Resolution X-ray Structural Studies of the Methanol Dehydrogenase from Methylococcus capsulatus (Bath).
J.Am.Chem.Soc., 143, 2021
|
|
1DAZ
 
 | | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease | | Descriptor: | HIV-1 PROTEASE (RETROPEPSIN), N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide | | Authors: | Mahalingam, B, Louis, J.M, Reed, C.C, Adomat, J.M, Krouse, J, Wang, Y.F, Harrison, R.W, Weber, I.T. | | Deposit date: | 1999-11-01 | | Release date: | 2000-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease.
Eur.J.Biochem., 263, 1999
|
|
1DKU
 
 | | CRYSTAL STRUCTURES OF BACILLUS SUBTILIS PHOSPHORIBOSYLPYROPHOSPHATE SYNTHETASE: MOLECULAR BASIS OF ALLOSTERIC INHIBITION AND ACTIVATION. | | Descriptor: | METHYL PHOSPHONIC ACID ADENOSINE ESTER, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, PROTEIN (PHOSPHORIBOSYL PYROPHOSPHATE SYNTHETASE) | | Authors: | Eriksen, T.A, Kadziola, A, Bentsen, A.-K, Harlow, K.W, Larsen, S. | | Deposit date: | 1999-12-08 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the function of Bacillus subtilis phosphoribosyl-pyrophosphate synthetase.
Nat.Struct.Biol., 7, 2000
|
|
7CFX
 
 | | NAD-soaked Holo-methanol dehydrogenase (MDH) from Methylococcus capsulatus (Bath) | | Descriptor: | CALCIUM ION, Methanol dehydrogenase [cytochrome c] subunit 2, Methanol dehydrogenase protein, ... | | Authors: | Chuankhayan, P, Chan, S.I, Nareddy, P.K.R, Tsai, I.K, Tsai, Y.F, Chen, K.H.-C, Yu, S.S.-F, Chen, C.J. | | Deposit date: | 2020-06-29 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of Pyrroloquinoline Quinone-Dependent Hydride Transfer Chemistry from Spectroscopic and High-Resolution X-ray Structural Studies of the Methanol Dehydrogenase from Methylococcus capsulatus (Bath).
J.Am.Chem.Soc., 143, 2021
|
|
1DBG
 
 | | CRYSTAL STRUCTURE OF CHONDROITINASE B | | Descriptor: | 4-deoxy-alpha-D-glucopyranose-(1-3)-[beta-D-glucopyranose-(1-4)]2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, CHONDROITINASE B | | Authors: | Huang, W, Matte, A, Li, Y, Kim, Y.S, Linhardt, R.J, Su, H, Cygler, M. | | Deposit date: | 1999-11-02 | | Release date: | 2000-01-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of chondroitinase B from Flavobacterium heparinum and its complex with a disaccharide product at 1.7 A resolution.
J.Mol.Biol., 294, 1999
|
|
1DLI
 
 | | THE FIRST STRUCTURE OF UDP-GLUCOSE DEHYDROGENASE (UDPGDH) REVEALS THE CATALYTIC RESIDUES NECESSARY FOR THE TWO-FOLD OXIDATION | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Campbell, R.E, Mosimann, S.C, van de Rijn, I, Tanner, M.E, Strynadka, N.C.J. | | Deposit date: | 1999-12-09 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The first structure of UDP-glucose dehydrogenase reveals the catalytic residues necessary for the two-fold oxidation.
Biochemistry, 39, 2000
|
|
1DLW
 
 | | X-RAY CRYSTAL STRUCTURE OF TRUNCATED HEMOGLOBIN FROM P.CAUDATUM. | | Descriptor: | HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pesce, A, Couture, M, Guertin, M, Dewilde, S, Moens, L, Bolognesi, M. | | Deposit date: | 1999-12-13 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A novel two-over-two alpha-helical sandwich fold is characteristic of the truncated hemoglobin family.
EMBO J., 19, 2000
|
|
1DCT
 
 | | DNA (CYTOSINE-5) METHYLASE FROM HAEIII COVALENTLY BOUND TO DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*CP*CP*AP*GP*CP*AP*GP*GP*(C49)P*CP*AP*CP*CP*AP*GP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*CP*TP*GP*GP*TP*GP*GP*(C5M)P*CP*TP*GP*CP*TP*GP*G)-3'), ... | | Authors: | Reinisch, K.M, Chen, L, Verdine, G.L, Lipscomb, W.N. | | Deposit date: | 1995-05-17 | | Release date: | 1995-09-15 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of HaeIII methyltransferase convalently complexed to DNA: an extrahelical cytosine and rearranged base pairing.
Cell(Cambridge,Mass.), 82, 1995
|
|
7CE5
 
 | | Methanol-PQQ bound methanol dehydrogenase (MDH) from Methylococcus capsulatus (Bath) | | Descriptor: | CALCIUM ION, METHANOL, Methanol dehydrogenase [cytochrome c] subunit 2, ... | | Authors: | Chuankhayan, P, Chan, S.I, Nareddy, P.K.R, Tsai, I.K, Tsai, Y.F, Chen, K.H.-C, Yu, S.S.-F, Chen, C.J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of Pyrroloquinoline Quinone-Dependent Hydride Transfer Chemistry from Spectroscopic and High-Resolution X-ray Structural Studies of the Methanol Dehydrogenase from Methylococcus capsulatus (Bath).
J.Am.Chem.Soc., 143, 2021
|
|
1DNC
 
 | | HUMAN GLUTATHIONE REDUCTASE MODIFIED BY DIGLUTATHIONE-DINITROSO-IRON | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE, GLUTATHIONE REDUCTASE, ... | | Authors: | Becker, K, Savvides, S.N, Keese, M, Schirmer, R.H, Karplus, P.A. | | Deposit date: | 1998-02-20 | | Release date: | 1998-05-27 | | Last modified: | 2011-12-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enzyme inactivation through sulfhydryl oxidation by physiologic NO-carriers.
Nat.Struct.Biol., 5, 1998
|
|
1DE8
 
 | | HUMAN APURINIC/APYRIMIDINIC ENDONUCLEASE-1 (APE1) BOUND TO ABASIC DNA | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*GP*GP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*CP*(3DR)P*GP*AP*TP*CP*G)-3'), MAJOR APURINIC/APYRIMIDINIC ENDONUCLEASE | | Authors: | Mol, C.D, Izumi, T, Mitra, S, Tainer, J.A. | | Deposit date: | 1999-11-13 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected
Nature, 403, 2000
|
|
7CFU
 
 | |
5I84
 
 | |
1DHK
 
 | | STRUCTURE OF PORCINE PANCREATIC ALPHA-AMYLASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BEAN LECTIN-LIKE INHIBITOR, ... | | Authors: | Bompard-Gilles, C, Payan, F. | | Deposit date: | 1996-10-14 | | Release date: | 1997-12-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate mimicry in the active center of a mammalian alpha-amylase: structural analysis of an enzyme-inhibitor complex.
Structure, 4, 1996
|
|
1DVR
 
 | |
5IBB
 
 | | Structure of T. thermophilus 70S ribosome complex with mRNA, tRNAfMet and cognate tRNAVal in the A-site | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2016-02-22 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The ribosome prohibits the GU wobble geometry at the first position of the codon-anticodon helix.
Nucleic Acids Res., 44, 2016
|
|
1DBM
 
 | |
1DIL
 
 | | SIALIDASE FROM SALMONELLA TYPHIMURIUM COMPLEXED WITH APANA AND EPANA INHIBITORS | | Descriptor: | (1R)-4-acetamido-1,5-anhydro-2,4-dideoxy-1-phosphono-D-glycero-D-galacto-octitol, (1S)-4-acetamido-1,5-anhydro-2,4-dideoxy-1-phosphono-D-glycero-D-galacto-octitol, POTASSIUM ION, ... | | Authors: | Garman, E.F, Crennell, S.C, Vimr, E.R, Laver, W.G, Taylor, G.L. | | Deposit date: | 1996-04-23 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structures of Salmonella typhimurium LT2 neuraminidase and its complexes with three inhibitors at high resolution.
J.Mol.Biol., 259, 1996
|
|
1DLA
 
 | | NOVEL NADPH-BINDING DOMAIN REVEALED BY THE CRYSTAL STRUCTURE OF ALDOSE REDUCTASE | | Descriptor: | ALDOSE REDUCTASE | | Authors: | Rondeau, J.-M, Tete-Favier, F, Podjarny, A, Reymann, J.-M, Barth, P, Biellmann, J.-F, Moras, D. | | Deposit date: | 1993-02-08 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Novel NADPH-binding domain revealed by the crystal structure of aldose reductase.
Nature, 355, 1992
|
|
1DOR
 
 | | DIHYDROOROTATE DEHYDROGENASE A FROM LACTOCOCCUS LACTIS | | Descriptor: | DIHYDROOROTATE DEHYDROGENASE A, FLAVIN MONONUCLEOTIDE | | Authors: | Rowland, P, Larsen, S. | | Deposit date: | 1997-01-14 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the flavin containing enzyme dihydroorotate dehydrogenase A from Lactococcus lactis.
Structure, 5, 1997
|
|
