6UTB
 
 | |
5GJG
 
 | | Crystal structure of human TAK1/TAB1 fusion protein in complex with ligand 4 | | Descriptor: | N-(2-isopropoxy-4-(4-methylpiperazine-1-carbonyl)phenyl)-2-(3-(phenylcarbamoyl)phenyl)thiazole-4-carboxamide, TAK1 kinase - TAB1 chimera fusion protein | | Authors: | Irie, M, Nakamura, M, Fukami, T.A, Matsuura, T, Morishima, K. | | Deposit date: | 2016-06-29 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Development of a Method for Converting a TAK1 Type I Inhibitor into a Type II or c-Helix-Out Inhibitor by Structure-Based Drug Design (SBDD)
Chem.Pharm.Bull., 64, 2016
|
|
2IWI
 
 | | CRYSTAL STRUCTURE OF THE HUMAN PIM2 IN COMPLEX WITH A RUTHENIUM ORGANOMETALLIC LIGAND RU1 | | Descriptor: | RUTHENIUM-PYRIDOCARBAZOLE-1, SERINE/THREONINE-PROTEIN KINASE PIM-2 | | Authors: | Russo, S, Debreczeni, J.E, Amos, A, Bullock, A.N, Fedorov, O, Niesen, F, Sobott, F, Turnbull, A, Pike, A.C.W, Ugochukwu, E, Papagrigoriou, E, Bunkoczi, G, Gorrec, F, Edwards, A, Arrowsmith, C, Weigelt, J, Sundstrom, M, von Delft, F, Knapp, S. | | Deposit date: | 2006-06-30 | | Release date: | 2006-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the PIM2 kinase in complex with an organoruthenium inhibitor.
PLoS ONE, 4, 2009
|
|
2IU0
 
 | | crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 1,5-DIHYDROIMIDAZO[4,5-C][1,2,6]THIADIAZIN-4(3H)-ONE 2,2-DIOXIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, POTASSIUM ION | | Authors: | Xu, L, Chong, Y, Hwang, I, Onofrio, A.D, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2006-05-26 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure-Based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase
J.Biol.Chem., 282, 2007
|
|
6EKE
 
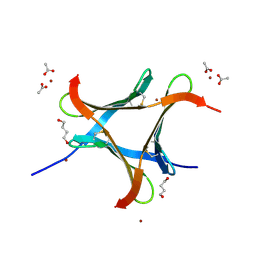 | |
5H3Q
 
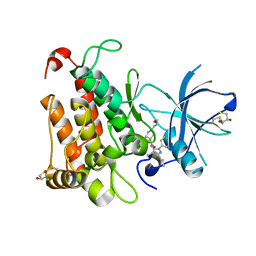 | | Crystal Structure of TrkA kinase with ligand | | Descriptor: | 1-[(3S,4R)-4-[3,4-bis(fluoranyl)phenyl]-1-(2-methoxyethyl)pyrrolidin-3-yl]-3-(5-ethoxy-4-methyl-2-phenyl-pyrazol-3-yl)urea, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, High affinity nerve growth factor receptor, ... | | Authors: | Noritaka, F. | | Deposit date: | 2016-10-26 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The juxtamembrane region of TrkA kinase is critical for inhibitor selectivity
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5K9G
 
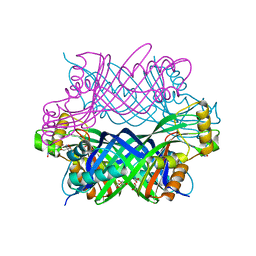 | | Crystal Structure of GTP Cyclohydrolase-IB with Tris | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, ... | | Authors: | Alvarez, J, Stec, B, Swairjo, M.A. | | Deposit date: | 2016-05-31 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism and catalytic strategy of the prokaryotic-specific GTP cyclohydrolase-IB.
Biochem.J., 474, 2017
|
|
5U4L
 
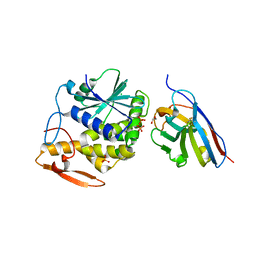 | | RTA-V1C7_G29R-high-salt | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Rudolph, M.J, Mantis, N. | | Deposit date: | 2016-12-05 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Using homology modeling to interrogate binding affinity in neutralization of ricin toxin by a family of single domain antibodies.
Proteins, 85, 2017
|
|
7JTH
 
 | |
7K14
 
 | | Ternary soak structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens with FMN and methanesulfonate | | Descriptor: | Alkanesulfonate monooxygenase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liew, J.J.M, Dowling, D.P, El Saudi, I.M. | | Deposit date: | 2020-09-07 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
5H32
 
 | | Cryo-EM structure of zika virus complexed with Fab C10 at pH 5.0 | | Descriptor: | C10 IgG heavy chain variable region, C10 IgG light chain variable region, structural protein E | | Authors: | Zhang, S, Kostyuchenko, V, Ng, T.-S, Lok, S.-M. | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Neutralization mechanism of a highly potent antibody against Zika virus
Nat Commun, 7, 2016
|
|
7K64
 
 | | Binary titrated soak structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens with FMN | | Descriptor: | Alkanesulfonate monooxygenase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liew, J.J.M, Dowling, D.P. | | Deposit date: | 2020-09-18 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
5GRJ
 
 | | Crystal structure of human PD-L1 with monoclonal antibody avelumab | | Descriptor: | Programmed cell death 1 ligand 1, avelumab H chain, avelumab L chain | | Authors: | Liu, K, Tan, S, Chai, Y, Chen, D, Song, H, Zhang, C.W.-H, Shi, Y, Liu, J, Tan, W, Lyu, J, Gao, S, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2016-08-11 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.206 Å) | | Cite: | Structural basis of anti-PD-L1 monoclonal antibody avelumab for tumor therapy.
Cell Res., 27, 2017
|
|
6ANJ
 
 | | Synaptotagmin-7, C2A domain | | Descriptor: | ACETATE ION, CALCIUM ION, Synaptotagmin-7, ... | | Authors: | Tomchick, D.R, Rizo, J, Voleti, R. | | Deposit date: | 2017-08-13 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Exceptionally tight membrane-binding may explain the key role of the synaptotagmin-7 C2A domain in asynchronous neurotransmitter release.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5Q6E
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 151) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QKO
 
 | |
1DQI
 
 | | CRYSTAL STRUCTURE OF SUPEROXIDE REDUCTASE FROM P. FURIOSUS IN THE OXIDIZED STATE AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | FE (III) ION, SUPEROXIDE REDUCTASE | | Authors: | Yeh, A.P, Hu, Y, Jenney Jr, F.E, Adams, M.W.W, Rees, D.C. | | Deposit date: | 2000-01-04 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the superoxide reductase from Pyrococcus furiosus in the oxidized and reduced states.
Biochemistry, 39, 2000
|
|
5Q6T
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 166) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q79
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 182) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QL3
 
 | |
5Q7P
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 198) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QLM
 
 | |
5Q80
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 209) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QM0
 
 | |
5Q8I
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 228) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
