5QY8
 
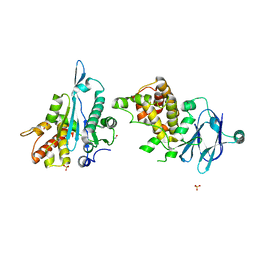 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment F2X-Entry C10a | | Descriptor: | 1,3-benzodioxole-5-carbothioamide, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
4FXV
 
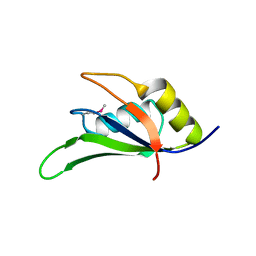 | |
3NDW
 
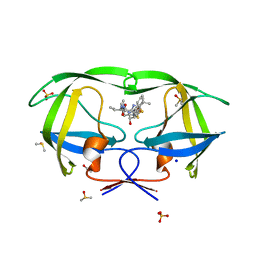 | | HIV-1 Protease Saquinavir:Ritonavir 1:15 complex structure | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Protease, ... | | Authors: | Geremia, S, Olajuyigbe, F.M, Demitri, N. | | Deposit date: | 2010-06-08 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Investigation of 2-Fold Disorder of Inhibitors and Relative Potency by Crystallizations of HIV-1 Protease in Ritonavir and Saquinavir Mixtures
Cryst.Growth Des., 11, 2011
|
|
4OP8
 
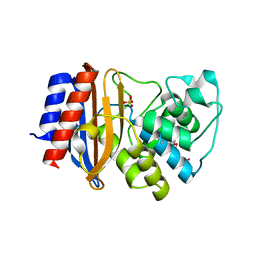 | | Room temperature crystal structure of stabilized TEM-1 beta-lactamase variant v.13 carrying G238S mutation | | Descriptor: | CALCIUM ION, SULFATE ION, TEM-94 ES-beta-lactamase, ... | | Authors: | Dellus-Gur, E, Elias, M, Fraser, J.S, Tawfik, D.S. | | Deposit date: | 2014-02-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Negative Epistasis and Evolvability in TEM-1 beta-Lactamase--The Thin Line between an Enzyme's Conformational Freedom and Disorder.
J. Mol. Biol., 427, 2015
|
|
4OPQ
 
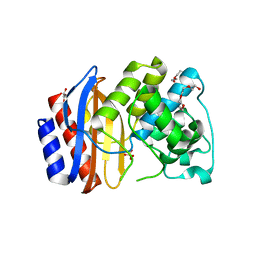 | | Room temperature crystal structure of stabilized TEM-1 beta-lactamase variant v.13 carrying R164S/G238S mutations | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Dellus-Gur, E, Elias, M, Fraser, J.S, Tawfik, D.S. | | Deposit date: | 2014-02-06 | | Release date: | 2015-05-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Negative Epistasis and Evolvability in TEM-1 beta-Lactamase--The Thin Line between an Enzyme's Conformational Freedom and Disorder.
J. Mol. Biol., 427, 2015
|
|
3NH8
 
 | | Crystal structure of murine aminoacylase 3 in complex with N-acetyl-S-1,2-dichlorovinyl-L-cysteine | | Descriptor: | Aspartoacylase-2, CHLORIDE ION, N-acetyl-S-[(1S)-1,2-dichloroethyl]-L-cysteine, ... | | Authors: | Hsieh, J.M, Tsirulnikov, K, Sawaya, M.R, Magilnick, N, Abuladze, N, Kurtz, I, Abramson, J, Pushkin, A. | | Deposit date: | 2010-06-14 | | Release date: | 2010-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structures of aminoacylase 3 in complex with acetylated substrates.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3UWC
 
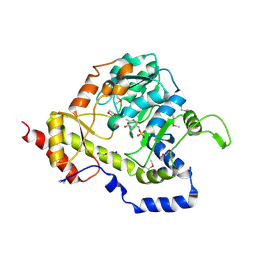 | | Structure of an aminotransferase (DegT-DnrJ-EryC1-StrS family) from Coxiella burnetii in complex with PMP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Cheung, J, Franklin, M, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-12-01 | | Release date: | 2011-12-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
2QMP
 
 | | Crystal Structure of HIV-1 protease complexed with PL-100 | | Descriptor: | N-[(5S)-5-{[(4-aminophenyl)sulfonyl](isobutyl)amino}-6-hydroxyhexyl]-Nalpha-(methoxycarbonyl)-beta-phenyl-L-phenylalaninamide, Pol polyprotein | | Authors: | Allison, T.J. | | Deposit date: | 2007-07-16 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of HIV-1 protease complexed with PL-100
To be Published
|
|
2WXV
 
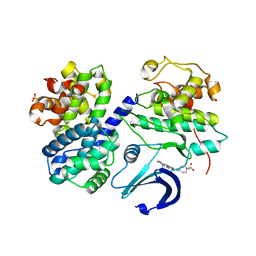 | | Structure of CDK2-CYCLIN A with a Pyrazolo(4,3-h) quinazoline-3- carboxamide inhibitor | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, N,1-DIMETHYL-8-{[1-(METHYLSULFONYL)PIPERIDIN-4-YL]AMINO}-1H-PYRAZOLO[4,3-H]QUINAZOLINE-3-CARBOXAMIDE, ... | | Authors: | Traquandi, G, Ciomei, M, Ballinari, D, Casale, E, Colombo, N, Croci, V, Fiorentini, F, Isacchi, A, Longo, A, Mercurio, C, Panzeri, A, Pastori, W, Pevarello, P, Volpi, D, Roussel, P, Vulpetti, A, Brasca, M.G. | | Deposit date: | 2009-11-10 | | Release date: | 2010-02-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of Potent Pyrazolo[4,3-H]Quinazoline-3-Carboxamides as Multi-Cyclin-Dependent Kinase Inhibitors.
J.Med.Chem., 53, 2010
|
|
2X8T
 
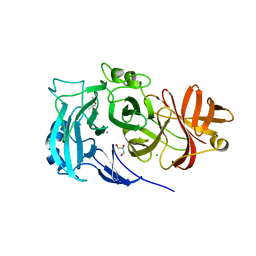 | | Crystal Structure of the Abn2 H318A mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | deSanctis, D, Inacio, J.M, Lindley, P.F, de Sa-Nogueira, I, Bento, I. | | Deposit date: | 2010-03-11 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | New Evidence for the Role of Calcium in the Glycosidase Reaction of Gh43 Arabinanases.
FEBS J., 277, 2010
|
|
5RER
 
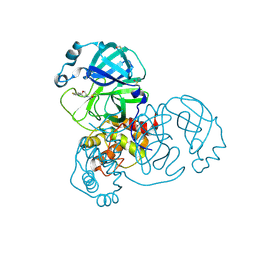 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102615 | | Descriptor: | 1-[(2R)-2-(4-fluorophenyl)morpholin-4-yl]ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
1L3E
 
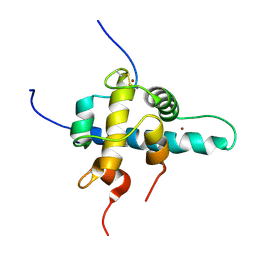 | | NMR Structures of the HIF-1alpha CTAD/p300 CH1 Complex | | Descriptor: | ZINC ION, hypoxia inducible factor-1 alpha subunit, p300 protein | | Authors: | Freedman, S.J, Sun, Z.J, Poy, F, Kung, A.L, Livingston, D.M, Wagner, G, Eck, M.J. | | Deposit date: | 2002-02-26 | | Release date: | 2002-04-24 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1UPP
 
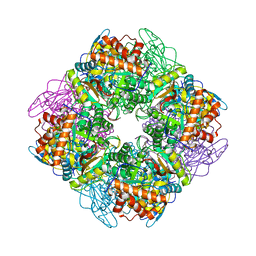 | | SPINACH RUBISCO IN COMPLEX WITH 2-CARBOXYARABINITOL 2 BISPHOSPHATE and Calcium. | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, CALCIUM ION, RIBULOSE BISPHOSPHATE CARBOXYLASE LARGE CHAIN, ... | | Authors: | Karkehabadi, S, Taylor, T.C, Andersson, I. | | Deposit date: | 2003-10-09 | | Release date: | 2003-10-14 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Calcium Supports Loop Closure But not Catalysis in Rubisco
J.Mol.Biol., 334, 2003
|
|
1UPM
 
 | | ACTIVATED SPINACH RUBISCO COMPLEXED WITH 2-CARBOXYARABINITOL 2 BISPHOSPHAT AND CA2+. | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, CALCIUM ION, RIBULOSE BISPHOSPHATE CARBOXYLASE LARGE CHAIN, ... | | Authors: | Karkehabadi, S, Taylor, T.C, Andersson, I. | | Deposit date: | 2003-10-08 | | Release date: | 2003-10-14 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Calcium Supports Loop Closure But not Catalysis in Rubisco
J.Mol.Biol., 334, 2003
|
|
4LFZ
 
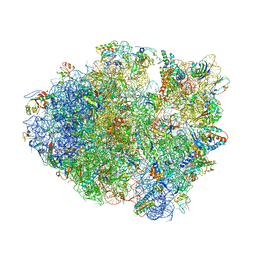 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 Bound to Codon CCC-U in the Absence of Paromomycin | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-06-27 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.92000055 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2CCY
 
 | | STRUCTURE OF FERRICYTOCHROME C(PRIME) FROM RHODOSPIRILLUM MOLISCHIANUM AT 1.67 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Finzel, B.C, Weber, P.C, Hardman, K.D, Salemme, F.R. | | Deposit date: | 1985-08-27 | | Release date: | 1986-01-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure of ferricytochrome c' from Rhodospirillum molischianum at 1.67 A resolution.
J.Mol.Biol., 186, 1985
|
|
1MD4
 
 | | A folding mutant of human class pi glutathione transferase, created by mutating glycine 146 of the wild-type protein to valine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, pi glutathione transferase | | Authors: | Kong, G.K.-W, Dragani, B, Aceto, A, Cocco, R, Mannervik, B, Stenberg, G, McKinstry, W.J, Polekhina, G, Parker, M.W. | | Deposit date: | 2002-08-06 | | Release date: | 2002-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Contribution of Glycine 146 to a Conserved Folding Module Affecting Stability and Refolding of Human Glutathione Transferase P1-1
J.Biol.Chem., 278, 2003
|
|
20GS
 
 | | GLUTATHIONE S-TRANSFERASE P1-1 COMPLEXED WITH CIBACRON BLUE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CIBACRON BLUE, GLUTATHIONE S-TRANSFERASE | | Authors: | Oakley, A.J, Lo Bello, M, Nuccetelli, M, Mazzetti, A.P, Parker, M.W. | | Deposit date: | 1997-12-16 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The ligandin (non-substrate) binding site of human Pi class glutathione transferase is located in the electrophile binding site (H-site).
J.Mol.Biol., 291, 1999
|
|
3V49
 
 | | Structure of ar lbd with activator peptide and sarm inhibitor 1 | | Descriptor: | 4-[(4R)-4-(4-hydroxyphenyl)-3,4-dimethyl-2,5-dioxoimidazolidin-1-yl]-2-(trifluoromethyl)benzonitrile, Androgen receptor, activator peptide, ... | | Authors: | Nique, F, Hebbe, S, Peixoto, C, Annoot, D, Lefrancois, J.-M, Duval, E, Michoux, L, Triballeau, N, Lemoullec, J.-M, Mollat, P, Thauvin, M, Prange, T, Minet, D, Clement-Lacroix, P, Robin-Jagerschmidt, C, Fleury, D, Guedin, D, Deprez, P. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of diarylhydantoins as new selective androgen receptor modulators.
J.Med.Chem., 55, 2012
|
|
3ELC
 
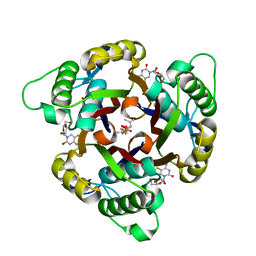 | | Crystal structure of 2C-methyl-D-erythritol 2,4-clycodiphosphate synthase complexed with ligand | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 4-amino-1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-5-fluoro-pyrimidin-2-one, GERANYL DIPHOSPHATE, ... | | Authors: | Hunter, W.N, Ramsden, N.L, Ulaganathan, V. | | Deposit date: | 2008-09-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structure-based approach to ligand discovery for 2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase: a target for antimicrobial therapy
J.Med.Chem., 52, 2009
|
|
1QJU
 
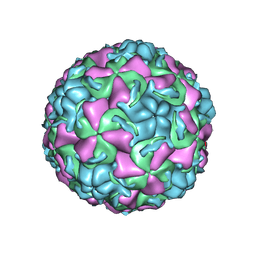 | | HUMAN RHINOVIRUS 16 COAT PROTEIN IN COMPLEX WITH ANTIVIRAL COMPOUND VP61209 | | Descriptor: | 2,6-DIMETHYL-1-(3-[3-METHYL-5-ISOXAZOLYL]-PROPANYL)-4-[2N-METHYL-2H-TETRAZOL-5-YL]-PHENOL, MYRISTIC ACID, PROTEIN VP1, ... | | Authors: | Hadfield, A.T, Minor, I, Diana, G.D, Rossmann, M.G. | | Deposit date: | 1999-07-05 | | Release date: | 1999-07-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of Three Structurally Related Antiviral Compounds in Complex with Human Rhinovirus 16
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2COJ
 
 | |
1ZTZ
 
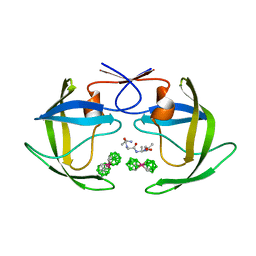 | | Crystal structure of HIV protease- metallacarborane complex | | Descriptor: | COBALT BIS(1,2-DICARBOLLIDE), PROTEASE RETROPEPSIN, autoproteolytic tetrapeptide | | Authors: | Cigler, P, Kozisek, M, Rezacova, P, Brynda, J, Otwinowski, Z, Sedlacek, J, Bodem, J, Kraeusslich, H.-G, Kral, V, Konvalinka, J. | | Deposit date: | 2005-05-28 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | From nonpeptide toward noncarbon protease inhibitors: Metallacarboranes as specific and potent inhibitors of HIV protease
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1QJY
 
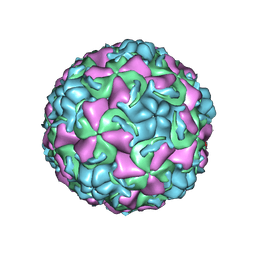 | | HUMAN RHINOVIRUS 16 COAT PROTEIN IN COMPLEX WITH ANTIVIRAL COMPOUND VP65099 | | Descriptor: | 2,6-DIMETHYL-1-(3-[3-METHYL-5-ISOXAZOLYL]-PROPANYL)-4-[2-METHYL-4-ISOXAZOLYL]-PHENOL, MYRISTIC ACID, PROTEIN VP1, ... | | Authors: | Hadfield, A.T, Diana, G.D, Rossmann, M.G. | | Deposit date: | 1999-07-06 | | Release date: | 1999-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of Three Structurally Related Antiviral Compounds in Complex with Human Rhinovirus 16
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
4HNO
 
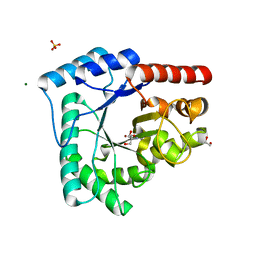 | | High resolution crystal structure of DNA Apurinic/apyrimidinic (AP) endonuclease IV Nfo from Thermatoga maritima | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Shin, D.S, Hosfield, D.J, Arvai, A.S, Tsutakawa, S.E, Tainer, J.A. | | Deposit date: | 2012-10-20 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.9194 Å) | | Cite: | Conserved Structural Chemistry for Incision Activity in Structurally Non-homologous Apurinic/Apyrimidinic Endonuclease APE1 and Endonuclease IV DNA Repair Enzymes.
J.Biol.Chem., 288, 2013
|
|
