1CIT
 
 | | DNA-BINDING MECHANISM OF THE MONOMERIC ORPHAN NUCLEAR RECEPTOR NGFI-B | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*AP*AP*AP*GP*GP*TP*CP*AP*TP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*AP*TP*GP*AP*CP*CP*TP*TP*TP*TP*CP*GP*G)-3'), PROTEIN (ORPHAN NUCLEAR RECEPTOR NGFI-B), ... | | Authors: | Meinke, G, Sigler, P.B. | | Deposit date: | 1999-04-05 | | Release date: | 1999-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | DNA-binding mechanism of the monomeric orphan nuclear receptor NGFI-B.
Nat.Struct.Biol., 6, 1999
|
|
1CZO
 
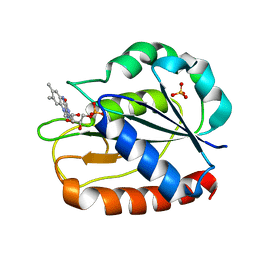 | | COMPARISONS OF WILD TYPE AND MUTANT FLAVODOXINS FROM ANACYSTIS NIDULANS. STRUCTURAL DETERMINANTS OF THE REDOX POTENTIALS. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, SULFATE ION | | Authors: | Hoover, D.M, Drennan, C.L, Metzger, A.L, Osborne, C, Weber, C.H, Pattridge, K.A, Ludwig, M.L. | | Deposit date: | 1999-09-03 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Comparisons of wild-type and mutant flavodoxins from Anacystis nidulans. Structural determinants of the redox potentials.
J.Mol.Biol., 294, 1999
|
|
5HKV
 
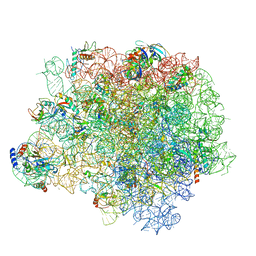 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus in complex with lincomycin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23s ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Yonath, A, Matzov, D, Eyal, Z, Ben Hamou, R, Zimmerman, E, Rozenberg, H, Bashan, A, Fridman, M. | | Deposit date: | 2016-01-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.66 Å) | | Cite: | Structural insights of lincosamides targeting the ribosome of Staphylococcus aureus.
Nucleic Acids Res., 45, 2017
|
|
1D02
 
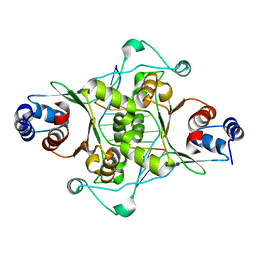 | | CRYSTAL STRUCTURE OF MUNI RESTRICTION ENDONUCLEASE IN COMPLEX WITH COGNATE DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*TP*TP*GP*GP*C)-3'), TYPE II RESTRICTION ENZYME MUNI | | Authors: | Deibert, M, Grazulis, S, Janulaitis, A, Siksnys, V, Huber, R. | | Deposit date: | 1999-09-08 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of MunI restriction endonuclease in complex with cognate DNA at 1.7 A resolution.
EMBO J., 18, 1999
|
|
1CJD
 
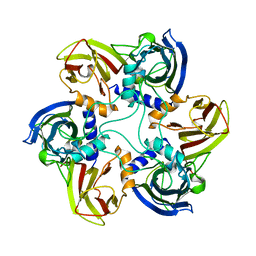 | | THE BACTERIOPHAGE PRD1 COAT PROTEIN, P3, IS STRUCTURALLY SIMILAR TO HUMAN ADENOVIRUS HEXON | | Descriptor: | PROTEIN (MAJOR CAPSID PROTEIN (P3)) | | Authors: | Benson, S.D, Bamford, J.K.H, Bamford, D.H, Burnett, R.M. | | Deposit date: | 1999-04-12 | | Release date: | 1999-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures.
Cell(Cambridge,Mass.), 98, 1999
|
|
1CK6
 
 | | BINDING MODE OF SALICYLHYDROXAMIC ACID TO ARTHROMYCES RAMOSUS PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PROTEIN (PEROXIDASE), ... | | Authors: | Fukuyama, K, Itakura, H. | | Deposit date: | 1999-04-28 | | Release date: | 1999-12-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding of salicylhydroxamic acid and several aromatic donor molecules to Arthromyces ramosus peroxidase, investigated by X-ray crystallography, optical difference spectroscopy, NMR relaxation, molecular dynamics, and kinetics.
Biochemistry, 38, 1999
|
|
1CPI
 
 | |
1D9E
 
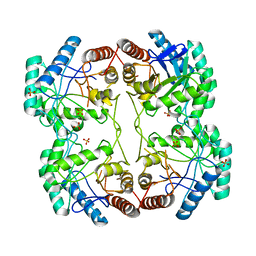 | | STRUCTURE OF E. COLI KDO8P SYNTHASE | | Descriptor: | 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE SYNTHASE, SULFATE ION | | Authors: | Radaev, S, Dastidar, P, Patel, M, Woodard, R.W, Gatti, D.L. | | Deposit date: | 1999-10-27 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of 3-deoxy-D-manno-octulosonate 8-phosphate synthase.
J.Biol.Chem., 275, 2000
|
|
1COM
 
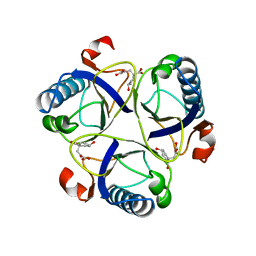 | | THE MONOFUNCTIONAL CHORISMATE MUTASE FROM BACILLUS SUBTILIS: STRUCTURE DETERMINATION OF CHORISMATE MUTASE AND ITS COMPLEXES WITH A TRANSITION STATE ANALOG AND PREPHENATE, AND IMPLICATIONS ON THE MECHANISM OF ENZYMATIC REACTION | | Descriptor: | CHORISMATE MUTASE, PREPHENIC ACID | | Authors: | Chook, Y.M, Ke, H, Lipscomb, W.N. | | Deposit date: | 1994-04-08 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The monofunctional chorismate mutase from Bacillus subtilis. Structure determination of chorismate mutase and its complexes with a transition state analog and prephenate, and implications for the mechanism of the enzymatic reaction.
J.Mol.Biol., 240, 1994
|
|
1CF8
 
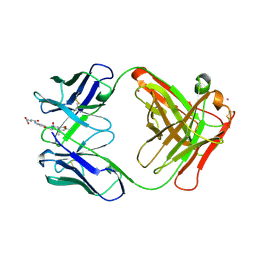 | | Convergence of catalytic antibody and terpene cyclase mechanisms: polyene cyclization directed by carbocation-pi interactions | | Descriptor: | 4-{4-[2-(1A,7A-DIMETHYL-4-OXY-OCTAHYDRO-1-OXA-4-AZA-CYCLOPROPA[A]NAPHTHALEN-4-YL) -ACETYLAMINO]-PHENYLCARBAMOYL}-BUTYRIC ACID, CADMIUM ION, PROTEIN (CATALYTIC ANTIBODY 19A4 (HEAVY CHAIN)), ... | | Authors: | Paschall, C.M, Hasserodt, J, Jones, T, Lerner, R.A, Janda, K.D, Christianson, D.W. | | Deposit date: | 1999-03-24 | | Release date: | 1999-07-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Convergence of Catalytic Antibody and Terpene Cyclase Mechanisms: Polyene Cyclization Directed by Carbocation-pi Interactions
Angew.Chem.Int.Ed.Engl., 38, 1999
|
|
1CPM
 
 | |
1CGE
 
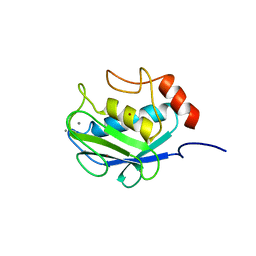 | | CRYSTAL STRUCTURES OF RECOMBINANT 19-KDA HUMAN FIBROBLAST COLLAGENASE COMPLEXED TO ITSELF | | Descriptor: | CALCIUM ION, FIBROBLAST COLLAGENASE, ZINC ION | | Authors: | Lovejoy, B, Hassell, A.M, Luther, M.A, Weigl, D, Jordan, S.R. | | Deposit date: | 1994-02-03 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of recombinant 19-kDa human fibroblast collagenase complexed to itself.
Biochemistry, 33, 1994
|
|
7BUG
 
 | | Reduced oxygenase of carbazole 1,9a-dioxygenase | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-METHOXYETHANOL, CARBONATE ION, ... | | Authors: | Matsuzawa, J, Wang, Y.X, Suzuki-Minakuchi, C, Nojiri, H. | | Deposit date: | 2020-04-06 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Reduced oxygenase of carbazole 1,9a-dioxygenase
To Be Published
|
|
1CQI
 
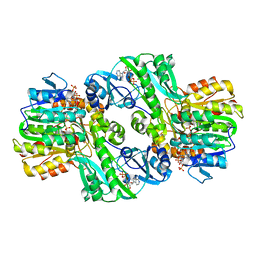 | | Crystal Structure of the Complex of ADP and MG2+ with Dephosphorylated E. Coli Succinyl-CoA Synthetase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, COENZYME A, MAGNESIUM ION, ... | | Authors: | Joyce, M.A, Fraser, M.E, James, M.N.G, Bridger, W.A, Wolodko, W.T. | | Deposit date: | 1999-08-06 | | Release date: | 2000-01-07 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | ADP-binding site of Escherichia coli succinyl-CoA synthetase revealed by x-ray crystallography.
Biochemistry, 39, 2000
|
|
1CRB
 
 | |
1CLO
 
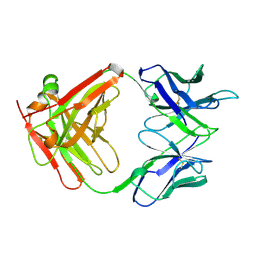 | |
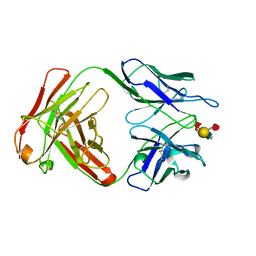
1CLY
 
 | |
1CRX
 
 | | CRE RECOMBINASE/DNA COMPLEX REACTION INTERMEDIATE I | | Descriptor: | CRE RECOMBINASE, DNA (5'-D(*AP*TP*AP*TP*GP*CP*TP*AP*TP*AP*CP*GP*AP*AP*GP*TP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*CP*TP*TP*CP*GP*TP*AP*TP*AP*G)-3'), ... | | Authors: | Guo, F, Gopaul, D.N, Van Duyne, G.D. | | Deposit date: | 1997-07-02 | | Release date: | 1998-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse.
Nature, 389, 1997
|
|
1CMX
 
 | |
5NUE
 
 | | Cytosolic Malate Dehydrogenase 1 (peroxide-treated) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-ETHOXYETHANOL, ... | | Authors: | Young, D, Messens, J, Huang, J, Reichheld, J.-P. | | Deposit date: | 2017-04-29 | | Release date: | 2018-02-28 | | Last modified: | 2018-07-04 | | Method: | X-RAY DIFFRACTION (1.35000277 Å) | | Cite: | Self-protection of cytosolic malate dehydrogenase against oxidative stress in Arabidopsis.
J. Exp. Bot., 69, 2018
|
|
1CTU
 
 | | TRANSITION-STATE SELECTIVITY FOR A SINGLE OH GROUP DURING CATALYSIS BY CYTIDINE DEAMINASE | | Descriptor: | 4-HYDROXY-3,4-DIHYDRO-ZEBULARINE, CYTIDINE DEAMINASE, ZINC ION | | Authors: | Xiang, S, Short, S.A, Wolfenden, R, Carter, C.W. | | Deposit date: | 1995-02-11 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Transition-state selectivity for a single hydroxyl group during catalysis by cytidine deaminase.
Biochemistry, 34, 1995
|
|
1CQN
 
 | |
5O5K
 
 | | X-ray structure of a bacterial adenylyl cyclase soluble domain | | Descriptor: | 3'-O-(N-METHYLANTHRANILOYL)-GUANOSINE-5'-TRIPHOSPHATE, Adenylate cyclase, MANGANESE (II) ION, ... | | Authors: | Vercellino, I, Korkhov, V.M. | | Deposit date: | 2017-06-02 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Role of the nucleotidyl cyclase helical domain in catalytically active dimer formation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1CR7
 
 | | PEANUT LECTIN-LACTOSE COMPLEX MONOCLINIC FORM | | Descriptor: | CALCIUM ION, LECTIN, MANGANESE (II) ION, ... | | Authors: | Ravishankar, R, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 1999-08-14 | | Release date: | 2001-04-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the peanut lectin-lactose complex at acidic pH: retention of unusual quaternary structure, empty and carbohydrate bound combining sites, molecular mimicry and crystal packing directed by interactions at the combining site.
Proteins, 43, 2001
|
|
1CVY
 
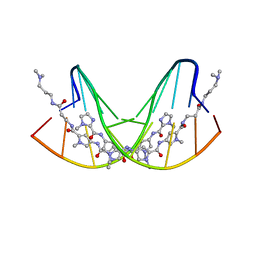 | | CRYSTAL STRUCTURE OF POLYAMIDE DIMER (IMPYPYPYBETADP)2 BOUND TO CCAGATCTGG | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*TP*CP*TP*GP*G)-3', IMIDAZOLE-PYRROLE POLYAMIDE | | Authors: | Kielkopf, C.L, Bremer, R.E, White, S, Baird, E.E, Dervan, P.B, Rees, D.C. | | Deposit date: | 1999-08-24 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural effects of DNA sequence on T.A recognition by hydroxypyrrole/pyrrole pairs in the minor groove.
J.Mol.Biol., 295, 2000
|
|
