7NPF
 
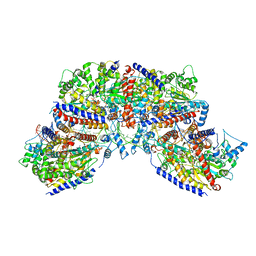 | | Vibrio cholerae ParA2-ATPyS-DNA filament | | Descriptor: | AAA family ATPase, DNA (49-MER), MAGNESIUM ION, ... | | Authors: | Parker, A.V, Bergeron, J.R.C. | | Deposit date: | 2021-02-26 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The structure of the bacterial DNA segregation ATPase filament reveals the conformational plasticity of ParA upon DNA binding.
Nat Commun, 12, 2021
|
|
1Y1Y
 
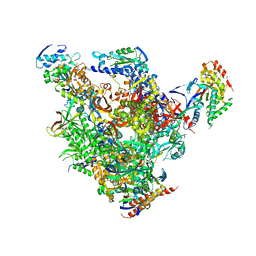 | | RNA Polymerase II-TFIIS-DNA/RNA complex | | Descriptor: | 5'-D(P*TP*AP*CP*GP*CP*CP*T)-3', 5'-R(P*AP*GP*GP*C)-3', DNA-directed RNA polymerase II 13.6 kDa polypeptide, ... | | Authors: | Cramer, P, Kettenberger, H, Armache, K.-J. | | Deposit date: | 2004-11-19 | | Release date: | 2004-12-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS.
Mol.Cell, 16, 2004
|
|
1Y1W
 
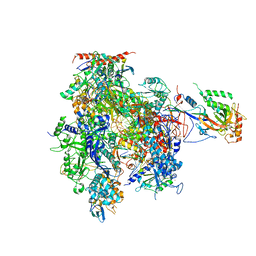 | | Complete RNA Polymerase II elongation complex | | Descriptor: | 5'-D(*AP*AP*GP*TP*AP*CP*T)-3', 5'-D(P*AP*GP*TP*AP*CP*TP*TP*AP*CP*GP*CP*CP*TP*GP*GP*TP*CP*AP*T)-3', 5'-R(*AP*AP*GP*AP*CP*CP*AP*GP*GP*C)-3', ... | | Authors: | Cramer, P, Kettenberger, H, Armache, K.-J. | | Deposit date: | 2004-11-19 | | Release date: | 2005-01-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Complete RNA Polymerase II Elongation Complex Structure and Its Interactions with NTP and TFIIS
Mol.Cell, 16, 2004
|
|
6VBD
 
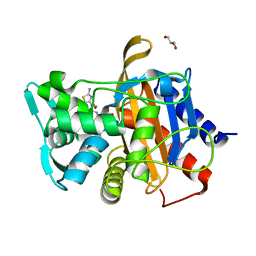 | |
6VBL
 
 | |
1Z8J
 
 | | Crystal structure of the thrombin mutant G193P bound to PPACK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, ... | | Authors: | Bobofchak, K.M, Pineda, A.O, Mathews, F.S, Di Cera, E. | | Deposit date: | 2005-03-30 | | Release date: | 2005-05-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Energetic and structural consequences of perturbing Gly-193 in the oxyanion hole of serine proteases
J.Biol.Chem., 280, 2005
|
|
6YGA
 
 | | Crystal structure of the apo NatC complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Grunwald, S, Hopf, L, Bock-Bierbaum, T, Lally, C.C, Spahn, C.M.T, Daumke, O. | | Deposit date: | 2020-03-27 | | Release date: | 2020-10-28 | | Last modified: | 2020-11-18 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Divergent architecture of the heterotrimeric NatC complex explains N-terminal acetylation of cognate substrates.
Nat Commun, 11, 2020
|
|
6TY3
 
 | | FAK structure from single particle analysis of 2D crystals | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Acebron, I, Righetto, R, Biyani, N, Chami, M, Boskovic, J, Stahlberg, H, Lietha, D. | | Deposit date: | 2020-01-15 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.32 Å) | | Cite: | Structural basis of Focal Adhesion Kinase activation on lipid membranes.
Embo J., 39, 2020
|
|
6TY4
 
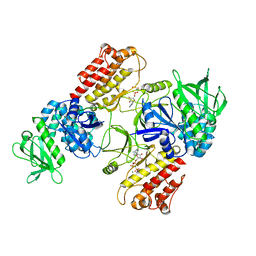 | | FAK structure with AMP-PNP from single particle analysis of 2D crystals | | Descriptor: | Focal adhesion kinase 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Acebron, I, Righetto, R, Biyani, N, Chami, M, Boskovic, J, Stahlberg, H, Lietha, D. | | Deposit date: | 2020-01-15 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.96 Å) | | Cite: | Structural basis of Focal Adhesion Kinase activation on lipid membranes.
Embo J., 39, 2020
|
|
5TPG
 
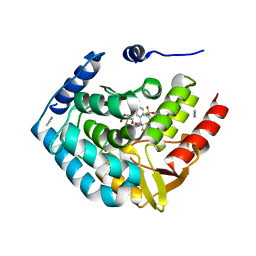 | | Optimization of spirocyclic proline tryptophanhydroxylase-1 inhibitors | | Descriptor: | (3S)-8-(2-amino-6-{(1R)-1-[5-chloro-3'-(methylsulfonyl)[1,1'-biphenyl]-2-yl]-2,2,2-trifluoroethoxy}pyrimidin-4-yl)-2,8-diazaspiro[4.5]decane-3-carboxylic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETONITRILE, ... | | Authors: | Stein, A.J, Goldberg, D.R, De Lombaert, S, Holt, M.C. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Optimization of spirocyclic proline tryptophan hydroxylase-1 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
4AZY
 
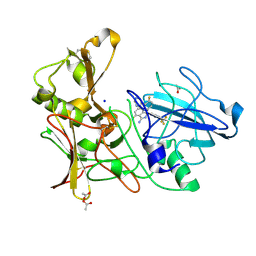 | | Design and Synthesis of BACE1 Inhibitors with In Vivo Brain Reduction of beta-Amyloid Peptides (COMPOUND 10) | | Descriptor: | (1S)-4-fluoro-1-(4-fluoro-3-pyrimidin-5-ylphenyl)-1-[2-(trifluoromethyl)pyridin-4-yl]-1H-isoindol-3-amine, ACETATE ION, BETA-SECRETASE 1, ... | | Authors: | Swahn, B.M, Kolmodin, K, Karlstrom, S, von Berg, S, Soderman, P, Holenz, J, Berg, S, Lindstrom, J, Sundstrom, M, Turek, D, Kihlstrom, J, Slivo, C, Andersson, L, Pyring, D, Ohberg, L, Kers, A, Bogar, K, Bergh, M, Olsson, L.L, Janson, J, Eketjall, S, Georgievska, B, Jeppsson, F, Falting, J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Design and synthesis of beta-site amyloid precursor protein cleaving enzyme (BACE1) inhibitors with in vivo brain reduction of beta-amyloid peptides.
J. Med. Chem., 55, 2012
|
|
6YGC
 
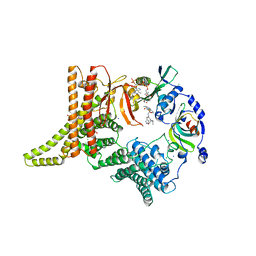 | | Crystal structure of the NatC complex bound to Arl3 peptide and CoA | | Descriptor: | ADP-ribosylation factor-like protein 3, CHLORIDE ION, COENZYME A, ... | | Authors: | Grunwald, S, Hopf, L, Bock-Bierbaum, T, Lally, C.C, Spahn, C.M.T, Daumke, O. | | Deposit date: | 2020-03-27 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | Divergent architecture of the heterotrimeric NatC complex explains N-terminal acetylation of cognate substrates.
Nat Commun, 11, 2020
|
|
6VEN
 
 | |
4B00
 
 | | Design and Synthesis of BACE1 Inhibitors with In Vivo Brain Reduction of beta-Amyloid Peptides (COMPOUND (R)-41) | | Descriptor: | 5-{(1R)-3-amino-4-fluoro-1-[3-(5-prop-1-yn-1-ylpyridin-3-yl)phenyl]-1H-isoindol-1-yl}-1-ethyl-3-methylpyridin-2(1H)-one, ACETATE ION, BETA-SECRETASE 1 | | Authors: | Swahn, B.M, Kolmodin, K, Karlstrom, S, von Berg, S, Soderman, P, Holenz, J, Berg, S, Lindstrom, J, Sundstrom, M, Turek, D, Kihlstrom, J, Slivo, C, Andersson, L, Pyring, D, Ohberg, L, Kers, A, Bogar, K, Bergh, M, Olsson, L.L, Janson, J, Eketjall, S, Georgievska, B, Jeppsson, F, Falting, J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Design and synthesis of beta-site amyloid precursor protein cleaving enzyme (BACE1) inhibitors with in vivo brain reduction of beta-amyloid peptides.
J. Med. Chem., 55, 2012
|
|
5VBC
 
 | | Crystal structure of ATXR5 in complex with histone H3.1 | | Descriptor: | DIMETHYL SULFOXIDE, Histone H3.1 peptide, Probable Histone-lysine N-methyltransferase ATXR5, ... | | Authors: | Couture, J.-F, Bergamin, E. | | Deposit date: | 2017-03-29 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
7L64
 
 | |
7L65
 
 | |
7L68
 
 | |
7L61
 
 | |
7L67
 
 | |
4HPG
 
 | | Crystal structure of a glycosylated beta-1,3-glucanase (HEV B 2), an allergen from Hevea brasiliensis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-1,3-glucanase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rodriguez-Romero, A, Hernandez-Santoyo, A. | | Deposit date: | 2012-10-23 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5364 Å) | | Cite: | Structural analysis of the endogenous glycoallergen Hev b 2 (endo-beta-1,3-glucanase) from Hevea brasiliensis and its recognition by human basophils.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6T3B
 
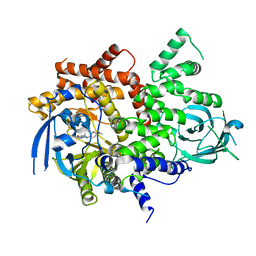 | | Crystal structure of PI3Kgamma with a dihydropurinone inhibitor (compound 4) | | Descriptor: | 2-[(4-methoxy-2-methyl-phenyl)amino]-7-methyl-9-(4-oxidanylcyclohexyl)purin-8-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Petersen, J, Oster, L, Schimpl, M, Goldberg, F.W, Finlay, M.R.V, Ting, A.K.T, Beattie, D, Lamont, G.M, Fallan, C, Wrigley, G.L, Howard, M.R, Williamson, B, Davies, B.R, Cadogan, E.B, Ramos-Montoya, A, Dean, E. | | Deposit date: | 2019-10-10 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | The Discovery of 7-Methyl-2-[(7-methyl[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(tetrahydro-2H-pyran-4-yl)-7,9-dihydro-8H-purin-8-one (AZD7648), a Potent and Selective DNA-Dependent Protein Kinase (DNA-PK) Inhibitor.
J.Med.Chem., 63, 2020
|
|
4DLG
 
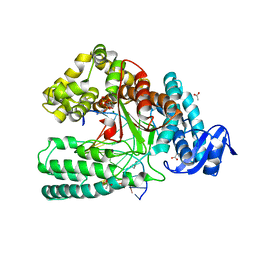 | | Ternary Structure of the large Fragment of Taq DNA polymerase | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, ACETATE ION, DNA polymerase I, ... | | Authors: | Marx, A, Diederichs, K, Obeid, S, Holzberger, B. | | Deposit date: | 2012-02-06 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural insights into the potential of 4-fluoroproline to modulate biophysical properties of protein
Chem Sci, 3, 2012
|
|
6SUW
 
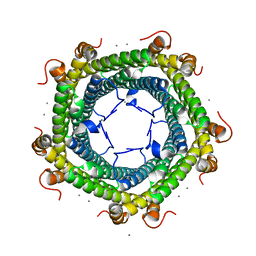 | |
4H25
 
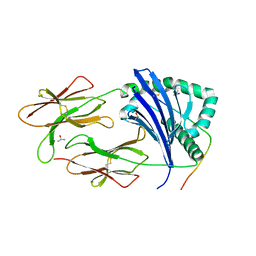 | | TCR interaction with peptide mimics of nickel offers structure insights to nickel contact allergy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Kappler, J.W, Yin, L, Dai, S, Marrack, P, Crawford, F. | | Deposit date: | 2012-09-12 | | Release date: | 2013-10-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | T-cell receptor (TCR) interaction with peptides that mimic nickel offers insight into nickel contact allergy.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
