2PTY
 
 | | Crystal Structure of the T. brucei enolase complexed with PEP | | Descriptor: | 1,2-ETHANEDIOL, Enolase, PHOSPHOENOLPYRUVATE, ... | | Authors: | Navarro, M.V.A.S, Rigden, D.J, Garratt, R.C, Dias, S.M.G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural flexibility in Trypanosoma brucei enolase revealed by X-ray crystallography and molecular dynamics.
Febs J., 274, 2007
|
|
1OO9
 
 | |
1U0C
 
 | | Y33C Mutant of Homing endonuclease I-CreI | | Descriptor: | 5'-D(*CP*GP*TP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*TP*AP*GP*C)-3', 5'-D(*GP*CP*TP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*AP*CP*G)-3', DNA endonuclease I-CreI, ... | | Authors: | Sussman, D, Chadsey, M, Fauce, S, Engel, A, Bruett, A, Monnat, R, Stoddard, B.L, Seligman, L.M. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Isolation and characterization of new homing endonuclease specificities at individual target site positions.
J.Mol.Biol., 342, 2004
|
|
1R6D
 
 | | Crystal Structure of DesIV double mutant (dTDP-glucose 4,6-dehydratase) from Streptomyces venezuelae with NAD and DAU bound | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TDP-glucose-4,6-dehydratase | | Authors: | Allard, S.T.M, Cleland, W.W, Holden, H.M. | | Deposit date: | 2003-10-14 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High Resolution X-ray Structure of dTDP-Glucose 4,6-Dehydratase from Streptomyces venezuelae
J.Biol.Chem., 279, 2004
|
|
1NSR
 
 | |
5W51
 
 | |
2YDU
 
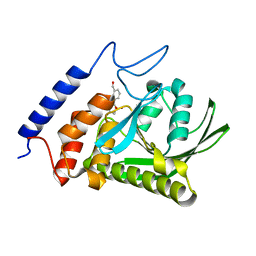 | | Crystal structure of YopH in complex with 3-(1,1-dioxido-3- oxoisothiazolidin-5-yl)benzaldeyde | | Descriptor: | 3-[(2S)-1,1-DIOXIDO-4-OXOTETRAHYDROTHIOPHEN-2-YL]BENZALDEHYDE, OUTER PROTEIN H PHOSPHATASE | | Authors: | Lountos, G.T, Kim, S.E, Bahta, M, Ulrich, R.G, Waugh, D.S, Burke, T.R. | | Deposit date: | 2011-03-24 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Isothiazolidinone (Izd) as a Phosphoryl Mimetic in Inhibitors of the Yersinia Pestis Protein Tyrosine Phosphatase Yoph.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2POW
 
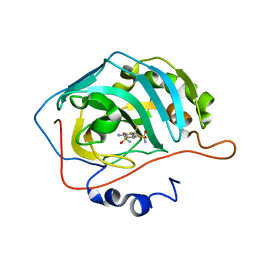 | | The crystal structure of the human carbonic anhydrase II in complex with 4-amino-6-trifluoromethyl-benzene-1,3-disulfonamide | | Descriptor: | 4-AMINO-6-(TRIFLUOROMETHYL)BENZENE-1,3-DISULFONAMIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Alterio, V, De Simone, G. | | Deposit date: | 2007-04-27 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Carbonic anhydrase inhibitors: Inhibition of human, bacterial, and archaeal isozymes with benzene-1,3-disulfonamides-Solution and crystallographic studies.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2FZE
 
 | |
2VK1
 
 | | Crystal structure of the Saccharomyces cerevisiae pyruvate decarboxylase variant D28A in complex with its substrate | | Descriptor: | MAGNESIUM ION, PYRUVATE DECARBOXYLASE ISOZYME 1, PYRUVIC ACID, ... | | Authors: | Kutter, S, Weik, M, Weiss, M.S, Konig, S. | | Deposit date: | 2007-12-16 | | Release date: | 2009-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Covalently Bound Substrate at the Regulatory Site of Yeast Pyruvate Decarboxylases Triggers Allosteric Enzyme Activation.
J.Biol.Chem., 284, 2009
|
|
1NSM
 
 | |
1NSZ
 
 | |
4HZO
 
 | | The Structure of the Bifunctional Acetyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double Hot Dog Fold | | Descriptor: | Bifunctional methylmalonyl-CoA:ACP acyltransferase/decarboxylase, CHLORIDE ION, COENZYME A | | Authors: | Lohman, J.R, Bingman, C.A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-15 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of the Bifunctional Acyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double-Hot-Dog Fold.
Biochemistry, 52, 2013
|
|
1TF4
 
 | | ENDO/EXOCELLULASE FROM THERMOMONOSPORA | | Descriptor: | CALCIUM ION, T. FUSCA ENDO/EXO-CELLULASE E4 CATALYTIC DOMAIN AND CELLULOSE-BINDING DOMAIN | | Authors: | Sakon, J, Wilson, D.B, Karplus, P.A. | | Deposit date: | 1997-05-30 | | Release date: | 1997-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca.
Nat.Struct.Biol., 4, 1997
|
|
4H8U
 
 | |
1Z5G
 
 | | Crystal structure of Salmonella typhimurium AphA protein | | Descriptor: | AphA protein, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Makde, R.D, Kumar, V. | | Deposit date: | 2005-03-18 | | Release date: | 2006-03-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and mutational analyses reveal the functional role of active-site Lys-154 and Asp-173 of Salmonella typhimurium AphA protein.
Arch.Biochem.Biophys., 464, 2007
|
|
1ORO
 
 | | A FLEXIBLE LOOP AT THE DIMER INTERFACE IS A PART OF THE ACTIVE SITE OF THE ADJACENT MONOMER OF ESCHERICHIA COLI OROTATE PHOSPHORIBOSYLTRANSFERASE | | Descriptor: | OROTATE PHOSPHORIBOSYLTRANSFERASE, SULFATE ION | | Authors: | Henriksen, A, Aghajari, N, Jensen, K.F, Gajhede, M. | | Deposit date: | 1995-09-11 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A flexible loop at the dimer interface is a part of the active site of the adjacent monomer of Escherichia coli orotate phosphoribosyltransferase.
Biochemistry, 35, 1996
|
|
1ZIP
 
 | | BACILLUS STEAROTHERMOPHILUS ADENYLATE KINASE | | Descriptor: | ADENYLATE KINASE, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Berry, M.B, Phillips Jr, G.N. | | Deposit date: | 1997-05-07 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of Bacillus stearothermophilus adenylate kinase with bound Ap5A, Mg2+ Ap5A, and Mn2+ Ap5A reveal an intermediate lid position and six coordinate octahedral geometry for bound Mg2+ and Mn2+.
Proteins, 32, 1998
|
|
3Q6K
 
 | | Salivary protein from Lutzomyia longipalpis | | Descriptor: | 43.2 kDa salivary protein, CITRIC ACID, SEROTONIN | | Authors: | Andersen, J.F, Xu, X, Chang, B.W, Collin, N, Valenzuela, J.G, Ribeiro, J.M. | | Deposit date: | 2011-01-02 | | Release date: | 2011-07-27 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure and function of a "yellow" protein from saliva of the sand fly Lutzomyia longipalpis that confers protective immunity against Leishmania major infection.
J.Biol.Chem., 286, 2011
|
|
1Z45
 
 | | Crystal structure of the gal10 fusion protein galactose mutarotase/UDP-galactose 4-epimerase from Saccharomyces cerevisiae complexed with NAD, UDP-glucose, and galactose | | Descriptor: | GAL10 bifunctional protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2005-03-15 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The molecular architecture of galactose mutarotase/UDP-galactose 4-epimerase from Saccharomyces cerevisiae.
J.Biol.Chem., 280, 2005
|
|
1XO1
 
 | | T5 5'-EXONUCLEASE MUTANT K83A | | Descriptor: | 5'-EXONUCLEASE | | Authors: | Ceska, T.A, Suck, D, Sayers, J.R. | | Deposit date: | 1998-11-19 | | Release date: | 1999-04-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis of conserved lysine residues in bacteriophage T5 5'-3' exonuclease suggests separate mechanisms of endo-and exonucleolytic cleavage.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1Z88
 
 | |
1P6B
 
 | | X-ray structure of phosphotriesterase, triple mutant H254G/H257W/L303T | | Descriptor: | DIETHYL 4-METHYLBENZYLPHOSPHONATE, ETHYL DIHYDROGEN PHOSPHATE, Parathion hydrolase, ... | | Authors: | Hill, C.M, Li, W, Thoden, J.B, Holden, H.M, Raushel, F.M. | | Deposit date: | 2003-04-29 | | Release date: | 2003-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Enhanced degradation of chemical warfare agents through molecular engineering of the phosphotriesterase active site.
J.Am.Chem.Soc., 125, 2003
|
|
1XVY
 
 | | Crystal Structure of iron-free Serratia marcescens SfuA | | Descriptor: | CITRIC ACID, sfuA | | Authors: | Shouldice, S.R, McRee, D.E, Dougan, D.R, Tari, L.W, Schryvers, A.B. | | Deposit date: | 2004-10-28 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Novel Anion-independent Iron Coordination by Members of a Third Class of Bacterial Periplasmic Ferric Ion-binding Proteins
J.Biol.Chem., 280, 2005
|
|
2HQM
 
 | |
