4IC0
 
 | | Crystal Structure of PAI-1 in Complex with Gallate | | Descriptor: | 3,4,5-trihydroxybenzoic acid, Plasminogen activator inhibitor 1 | | Authors: | Hong, Z.B, Lin, Z.H, Gong, L.H, Huang, M.D. | | Deposit date: | 2012-12-09 | | Release date: | 2013-12-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal Structure of PAI-1 in Complex with Gallate
To be Published
|
|
5R2D
 
 | | PanDDA analysis group deposition -- Endothiapepsin in complex with fragment F2X-Entry H11, DMSO-free | | Descriptor: | 1-cyclopentyl-3-[[(2~{S})-oxolan-2-yl]methyl]urea, Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.919 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
4DFW
 
 | | Oxime-based Post Solid-phase Peptide Diversification: Identification of High Affinity Polo-like Kinase 1 (Plk1) Polo-box Domain Binding Peptides | | Descriptor: | CHLORIDE ION, Peptide, Serine/threonine-protein kinase PLK1 | | Authors: | Liu, F, Park, J.-E, Qian, W.-J, Lim, D, Gr ber, M, Berg, T, Yaffe, M.B, Lee, K.S, Burke Jr, T.R. | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Oxime-based Post Solid-phase Peptide Diversification: Identification of High Affinity Polo-like Kinase 1 (Plk1) Polo-box Domain Binding Peptides
Chem.Biol., 2012
|
|
4IRE
 
 | | Crystal structure of GLIC with mutations at the loop C region | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ACETATE ION, OXALATE ION, ... | | Authors: | Chen, Q, Pan, J, Liang, Y.H, Xu, Y, Tang, P. | | Deposit date: | 2013-01-14 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Signal transduction pathways in the pentameric ligand-gated ion channels.
Plos One, 8, 2013
|
|
5R4F
 
 | | PanDDA analysis group deposition of ground-state model of ATAD2 | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Snee, M, Talon, R, Fowley, D, Collins, P, Nelson, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Von-Delft, F. | | Deposit date: | 2020-02-21 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
4ITX
 
 | | P113S mutant of E. coli Cystathionine beta-lyase MetC inhibited by reaction with L-Ala-P | | Descriptor: | CALCIUM ION, Cystathionine beta-lyase MetC, {1-[(3-HYDROXY-METHYL-5-PHOSPHONOOXY-METHYL-PYRIDIN-4-YLMETHYL)-AMINO]-ETHYL}-PHOSPHONIC ACID | | Authors: | Squire, C.J, Yosaatmadja, Y, Soo, V.W.C, Patrick, W.M. | | Deposit date: | 2013-01-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Mechanistic and Evolutionary Insights from the Reciprocal Promiscuity of Two Pyridoxal Phosphate-dependent Enzymes.
J.Biol.Chem., 291, 2016
|
|
3UIC
 
 | | Crystal Structure of FabI, an Enoyl Reductase from F. tularensis, in complex with a Novel and Potent Inhibitor | | Descriptor: | 1-(3,4-dichlorobenzyl)-5,6-dimethyl-1H-benzimidazole, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mehboob, S, Santarsiero, B.D, Truong, K, Johnson, M.E. | | Deposit date: | 2011-11-04 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and enzymatic analyses reveal the binding mode of a novel series of Francisella tularensis enoyl reductase (FabI) inhibitors.
J.Med.Chem., 55, 2012
|
|
2YMV
 
 | | Structure of Reduced M Smegmatis 5246, a homologue of M.Tuberculosis Acg | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, ACETATE ION, ACG NITROREDUCTASE, ... | | Authors: | Chauviac, F.-X, Bommer, M, Dobbek, H, Keep, N.H. | | Deposit date: | 2012-10-10 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Reduced Msacg, a Putative Nitroreductase from Mycobacterium Smegmatis and a Close Homologue of Mycobacterium Tuberculosis Acg.
J.Biol.Chem., 287, 2012
|
|
4R2R
 
 | | Wilms Tumor Protein (WT1) zinc fingers in complex with carboxylated DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(1CC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
2AN4
 
 | | Structure of PNMT complexed with S-adenosyl-L-homocysteine and the acceptor substrate octopamine | | Descriptor: | 4-(2R-AMINO-1-HYDROXYETHYL)PHENOL, PHOSPHATE ION, Phenylethanolamine N-methyltransferase, ... | | Authors: | Gee, C.L, Tyndall, J.D.A, Grunewald, G.L, Wu, Q, McLeish, M.J, Martin, J.L. | | Deposit date: | 2005-08-11 | | Release date: | 2006-03-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mode of binding of methyl acceptor substrates to the adrenaline-synthesizing enzyme phenylethanolamine N-methyltransferase: implications for catalysis
Biochemistry, 44, 2005
|
|
1UNE
 
 | |
3RL7
 
 | | Crystal structure of hDLG1-PDZ1 complexed with APC | | Descriptor: | 11-mer peptide from Adenomatous polyposis coli protein, Disks large homolog 1 | | Authors: | Zhang, Z, Li, H, Wu, G. | | Deposit date: | 2011-04-19 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the recognition of adenomatous polyposis coli by the Discs Large 1 protein.
Plos One, 6, 2011
|
|
2YEQ
 
 | | Structure of PhoD | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ALKALINE PHOSPHATASE D, ... | | Authors: | Lillington, J.E.D, Rodriguez, F, Roversi, P, Johnson, S.J, Berks, B, Lea, S.M. | | Deposit date: | 2011-03-30 | | Release date: | 2012-04-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of the Bacillus Subtilis Phosphodiesterase Phod Reveals an Iron and Calcium-Containing Active Site.
J.Biol.Chem., 289, 2014
|
|
4LP9
 
 | | Endothiapepsin complexed with Phe-reduced-Tyr peptide. | | Descriptor: | Endothiapepsin, GLYCEROL, SULFATE ION, ... | | Authors: | Guo, J, Cooper, J.B, Wood, S.P. | | Deposit date: | 2013-07-15 | | Release date: | 2014-01-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The structure of endothiapepsin complexed with a Phe-Tyr reduced-bond inhibitor at 1.35 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
1MR3
 
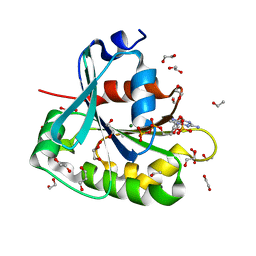 | | Saccharomyces cerevisiae ADP-ribosylation Factor 2 (ScArf2) complexed with GDP-3'P at 1.6A resolution | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ADP-ribosylation factor 2, ... | | Authors: | Amor, J.-C, Horton, J.R, Zhu, X, Wang, Y, Sullards, C, Ringe, D, Cheng, X, Kahn, R.A. | | Deposit date: | 2002-09-17 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of yeast ARF2 and ARL1: distinct roles for the N terminus in the structure and function of ARF family GTPases.
J.Biol.Chem., 276, 2001
|
|
4II2
 
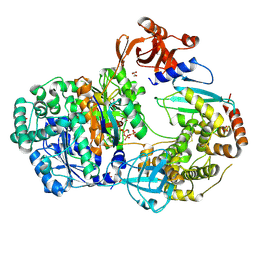 | | Crystal structure of Ubiquitin activating enzyme 1 (Uba1) in complex with the Ub E2 Ubc4, ubiquitin, and ATP/Mg | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Olsen, S.K, Lima, C.D. | | Deposit date: | 2012-12-19 | | Release date: | 2013-02-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a ubiquitin E1-E2 complex: insights to E1-E2 thioester transfer.
Mol.Cell, 49, 2013
|
|
7L2Y
 
 | |
2RE9
 
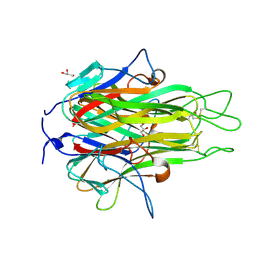 | | Crystal structure of TL1A at 2.1 A | | Descriptor: | GLYCEROL, MAGNESIUM ION, TNF superfamily ligand TL1A | | Authors: | Jin, T.C, Guo, F, Kim, S, Howard, A.J, Zhang, Y.Z. | | Deposit date: | 2007-09-25 | | Release date: | 2007-10-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of TNF ligand family member TL1A at 2.1 A.
Biochem.Biophys.Res.Commun., 364, 2007
|
|
4QG2
 
 | | Crystal structure of the tetrameric GTP/dATP/ATP-bound SAMHD1 (RN206) mutant catalytic core | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Koharudin, L.M.I, Wu, Y, DeLucia, M, Mehrens, J, Gronenborn, A.M, Ahn, J. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis of Allosteric Activation of Sterile alpha Motif and Histidine-Aspartate Domain-containing Protein 1 (SAMHD1) by Nucleoside Triphosphates.
J.Biol.Chem., 289, 2014
|
|
3RRV
 
 | |
2GTY
 
 | | Crystal structure of unliganded griffithsin | | Descriptor: | 1,2-ETHANEDIOL, Griffithsin, SULFATE ION | | Authors: | Ziolkowska, N.E, Wlodawer, A. | | Deposit date: | 2006-04-28 | | Release date: | 2006-08-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Domain-swapped structure of the potent antiviral protein griffithsin and its mode of carbohydrate binding.
Structure, 14, 2006
|
|
3O5V
 
 | | The Crystal Structure of the Creatinase/Prolidase N-terminal domain of an X-PRO dipeptidase from Streptococcus pyogenes to 1.85A | | Descriptor: | CHLORIDE ION, GLYCEROL, X-PRO dipeptidase | | Authors: | Stein, A.J, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of the Creatinase/Prolidase N-terminal domain of an X-PRO dipeptidase from Streptococcus pyogenes to 1.85A
To be Published
|
|
7LEC
 
 | | HIV-1 Protease WT (NL4-3) in Complex with PU3 (LR3-69) | | Descriptor: | Protease, SULFATE ION, diethyl ({4-[(2S,3R)-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxy-4-({[4-(hydroxymethyl)phenyl]sulfonyl}[(2S)-2-methylbutyl]amino)butyl]phenoxy}methyl)phosphonate | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2021-01-14 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | HIV-1 Protease Inhibitors with a P1 Phosphonate Modification Maintain Potency against Drug Resistant Variants by Increased van der Waals Contacts with Flaps Residues
To Be Published
|
|
1XSJ
 
 | | Structure of a Family 31 alpha glycosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Putative family 31 glucosidase yicI | | Authors: | Lovering, A.L, Lee, S.S, Kim, Y.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2004-10-19 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic and Structural Analysis of a Family 31 alpha-Glycosidase and Its Glycosyl-enzyme Intermediate
J.Biol.Chem., 280, 2005
|
|
4QG4
 
 | | Crystal structure of the tetrameric GTP/dATP/ATP-bound SAMHD1 (H210A) mutant catalytic core | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Koharudin, L.M.I, Wu, Y, DeLucia, M, Mehrens, J, Gronenborn, A.M, Ahn, J. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Allosteric Activation of Sterile alpha Motif and Histidine-Aspartate Domain-containing Protein 1 (SAMHD1) by Nucleoside Triphosphates.
J.Biol.Chem., 289, 2014
|
|
