5NJB
 
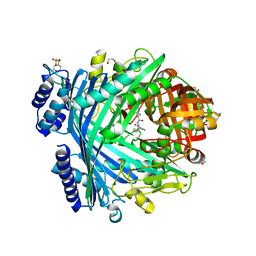 | | E. coli Microcin-processing metalloprotease TldD/E with actinonin bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACTINONIN, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5NJC
 
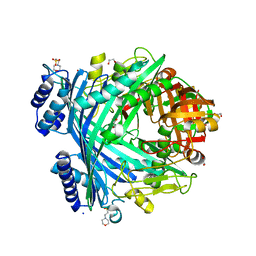 | | E. coli Microcin-processing metalloprotease TldD/E (TldD E263A mutant) with hexapeptide bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Metalloprotease PmbA, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5NHW
 
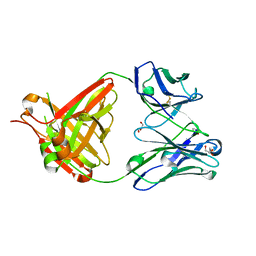 | | CRYSTAL STRUCTURE OF THE BIMAGRUMAB Fab | | Descriptor: | GLYCEROL, anti-human ActRII Bimagrumab Fab heavy-chain, anti-human ActRII Bimagrumab Fab light-chain | | Authors: | Rondeau, J.-M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Blockade of activin type II receptors with a dual anti-ActRIIA/IIB antibody is critical to promote maximal skeletal muscle hypertrophy.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5NJ5
 
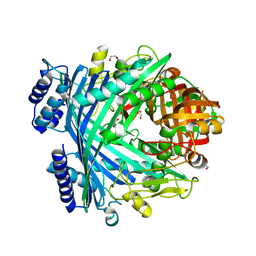 | | E. coli Microcin-processing metalloprotease TldD/E with phosphate bound | | Descriptor: | 1,2-ETHANEDIOL, Metalloprotease PmbA, Metalloprotease TldD, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5NHR
 
 | |
5NJ9
 
 | | E. coli Microcin-processing metalloprotease TldD/E with DRVY angiotensin fragment bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ASP-ARG-VAL-TYR, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
3UKW
 
 | | Mouse importin alpha: Bimax1 peptide complex | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bimax1 peptide, Importin subunit alpha-2 | | Authors: | Marfori, M, Forwood, J.K, Lonhienne, T.G, Kobe, B. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of High-Affinity Nuclear Localization Signal Interactions with Importin-alpha
Traffic, 13, 2012
|
|
3T7Z
 
 | | Structure of Methanocaldococcus jannaschii Nop N-terminal domain | | Descriptor: | ACETATE ION, GLYCEROL, Nucleolar protein Nop 56/58, ... | | Authors: | Biswas, S, Maxwell, S. | | Deposit date: | 2011-07-31 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structurally Conserved Nop56/58 N-terminal Domain Facilitates Archaeal Box C/D Ribonucleoprotein-guided Methyltransferase Activity.
J.Biol.Chem., 287, 2012
|
|
5GIP
 
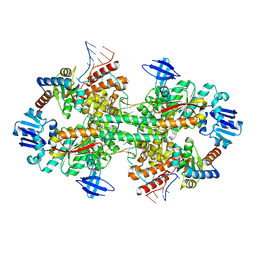 | | Crystal structure of box C/D RNP with 13 nt guide regions and 11 nt substrates | | Descriptor: | 50S ribosomal protein L7Ae, C/D RNA, C/D box methylation guide ribonucleoprotein complex aNOP56 subunit, ... | | Authors: | Yang, Z, Lin, J, Ye, K. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.129 Å) | | Cite: | Box C/D guide RNAs recognize a maximum of 10 nt of substrates
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3V9Y
 
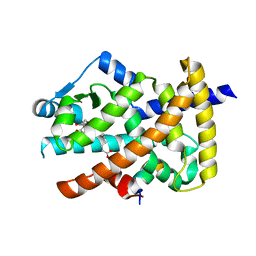 | | Crystal structure of the PPARgamma-LBD complexed with a cercosporamide derivative modulator | | Descriptor: | 4-{4-[({[(9aS)-8-acetyl-1,7-dihydroxy-3-methoxy-9a-methyl-9-oxo-9,9a-dihydrodibenzo[b,d]furan-4-yl]carbonyl}amino)methyl]naphthalen-2-yl}butanoic acid, Peptide from Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2011-12-28 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substituents at the naphthalene C3 position of (-)-Cercosporamide derivatives significantly affect the maximal efficacy as PPAR(gamma) partial agonists
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5NOF
 
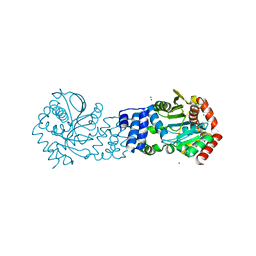 | | Anthranilate phosphoribosyltransferase from Thermococcus kodakaraensis | | Descriptor: | Anthranilate phosphoribosyltransferase, CHLORIDE ION, SODIUM ION, ... | | Authors: | Perveen, S, Rashid, N, Papageorgiou, A.C. | | Deposit date: | 2017-04-12 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Anthranilate phosphoribosyltransferase from the hyperthermophilic archaeon Thermococcus kodakarensis shows maximum activity with zinc and forms a unique dimeric structure.
FEBS Open Bio, 7, 2017
|
|
3PAF
 
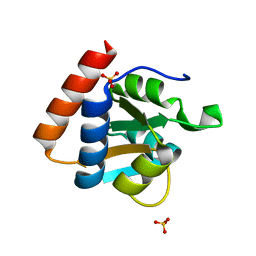 | | M. jannaschii L7Ae mutant | | Descriptor: | 50S ribosomal protein L7Ae, ACETATE ION, SULFATE ION | | Authors: | Biswas, S, Maxwell, E.S. | | Deposit date: | 2010-10-19 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and stability of M.jannaschii L7Ae El9 KtoQ mutant
To be Published
|
|
3O85
 
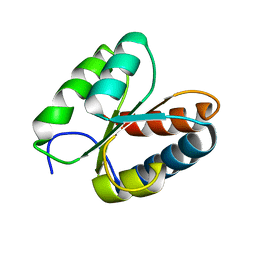 | | Giardia lamblia 15.5kD RNA binding protein | | Descriptor: | Ribosomal protein L7Ae | | Authors: | Biswas, S, Maxwell, E.S. | | Deposit date: | 2010-08-02 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.806 Å) | | Cite: | Comparative analysis of the 15.5kD box C/D snoRNP core protein in the primitive eukaryote Giardia lamblia reveals unique structural and functional features.
Biochemistry, 50, 2011
|
|
5NOE
 
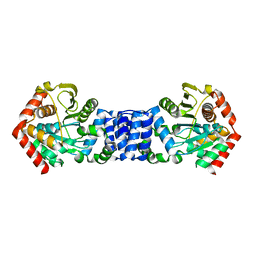 | |
5UI2
 
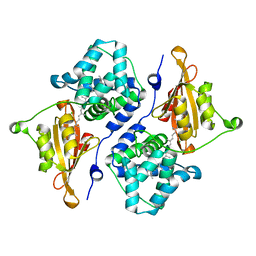 | | CRYSTAL STRUCTURE OF ORANGE CAROTENOID PROTEIN | | Descriptor: | (3'R)-3'-hydroxy-beta,beta-caroten-4-one, CHLORIDE ION, Orange carotenoid-binding protein, ... | | Authors: | KERFELD, C.A, SAWAYA, M.R, VISHNU, B, KROGMANN, D, YEATES, T.O. | | Deposit date: | 2017-01-12 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a cyanobacterial water-soluble carotenoid binding protein.
Structure, 11, 2003
|
|
6BRO
 
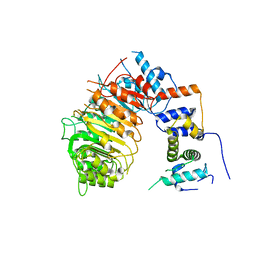 | |
6BRP
 
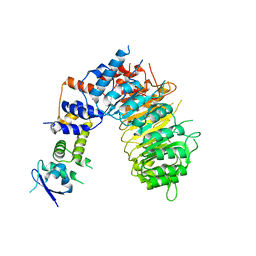 | | F-box protein form 2 | | Descriptor: | F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A | | Authors: | Shabek, N, Zheng, N, Mao, H, Hinds, T.R, Ticchiarelli, F, Leyser, O. | | Deposit date: | 2017-11-30 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling.
Nature, 563, 2018
|
|
6BRQ
 
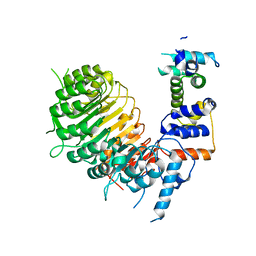 | | Crystal structure of rice ASK1-D3 ubiquitin ligase complex crystal form 3 | | Descriptor: | F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A | | Authors: | Shabek, N, Zheng, N, Mao, H, Hinds, T.R, Ticchiarelli, F, Leyser, O. | | Deposit date: | 2017-11-30 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling.
Nature, 563, 2018
|
|
9QZT
 
 | |
6TC7
 
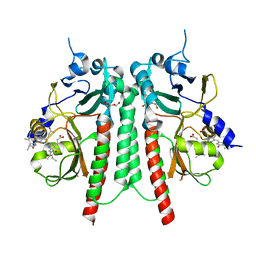 | | PAS-GAF bidomain of Glycine max phytochromeA | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHYCOCYANOBILIN, Phytochrome | | Authors: | Nagano, S, Guan, K, Shenkutie, S.M, Hughes, J.E. | | Deposit date: | 2019-11-05 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural insights into photoactivation and signalling in plant phytochromes.
Nat.Plants, 6, 2020
|
|
6TL4
 
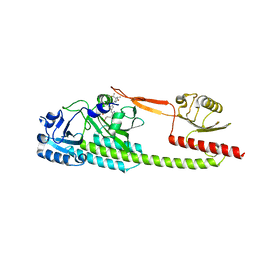 | | Photosensory module (PAS-GAF-PHY) of Glycine max phyB | | Descriptor: | PHYCOCYANOBILIN, Phytochrome | | Authors: | Nagano, S, Guan, K, Shenkutie, S.M, Hughes, J.E. | | Deposit date: | 2019-12-01 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into photoactivation and signalling in plant phytochromes.
Nat.Plants, 6, 2020
|
|
6UXI
 
 | | Structure of serine hydroxymethyltransferase 8 from Glycine max cultivar Essex complexed with PLP-Glycine | | Descriptor: | 1,2-ETHANEDIOL, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], Serine hydroxymethyltransferase | | Authors: | Korasick, D.A, Tanner, J.J, Beamer, L.J. | | Deposit date: | 2019-11-07 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Impaired folate binding of serine hydroxymethyltransferase 8 from soybean underlies resistance to the soybean cyst nematode.
J.Biol.Chem., 295, 2020
|
|
6UXK
 
 | |
6UXJ
 
 | | Structure of serine hydroxymethyltransferase 8 from Glycine max cultivar Essex complexed with PLP-glycine and 5-formyltetrahydrofolate | | Descriptor: | 1,2-ETHANEDIOL, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Korasick, D.A, Tanner, J.J, Beamer, L.J. | | Deposit date: | 2019-11-07 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Impaired folate binding of serine hydroxymethyltransferase 8 from soybean underlies resistance to the soybean cyst nematode.
J.Biol.Chem., 295, 2020
|
|
6UXH
 
 | |
