8STS
 
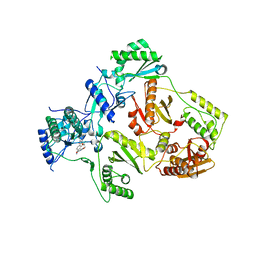 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C, V106A) varient in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-fluorophenoxy)-7-fluoro-2-naphthonitrile (JLJ636), a non-nucleoside inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-7-fluoronaphthalene-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Hollander, K, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Exploring novel HIV-1 reverse transcriptase inhibitors with drug-resistant mutants: A double mutant surprise.
Protein Sci., 32, 2023
|
|
8STQ
 
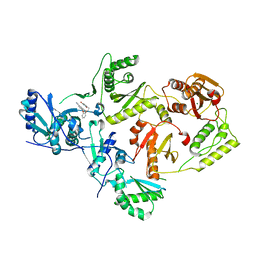 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) varient in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-2-naphthonitrile (JLJ600), a non-nucleoside inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}naphthalene-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Hollander, K, Frey, K.M, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | Exploring novel HIV-1 reverse transcriptase inhibitors with drug-resistant mutants: A double mutant surprise.
Protein Sci., 32, 2023
|
|
8STU
 
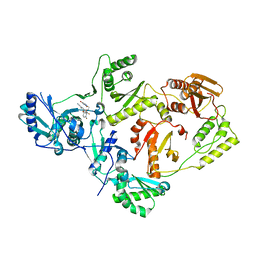 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C, V106A) variant in Complex with 8-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-fluorophenoxy)-6-fluoroindolizine-2-carbonitrile (JLJ578), a non-nucleoside inhibitor | | Descriptor: | 8-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-6-fluoroindolizine-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Hollander, K, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Exploring novel HIV-1 reverse transcriptase inhibitors with drug-resistant mutants: A double mutant surprise.
Protein Sci., 32, 2023
|
|
7Z7C
 
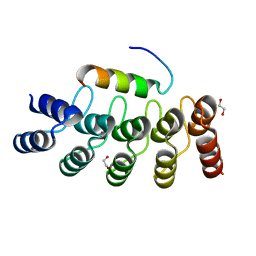 | |
7Z24
 
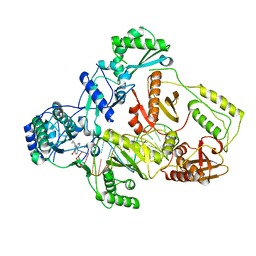 | | Cryo-EM structure of HIV-1 reverse transcriptase with a DNA aptamer in complex with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, DNA (38-mer), Reverse transcriptase/ribonuclease H | | Authors: | Singh, A.K, Das, K. | | Deposit date: | 2022-02-25 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-EM structures of wild-type and E138K/M184I mutant HIV-1 RT/DNA complexed with inhibitors doravirine and rilpivirine.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z29
 
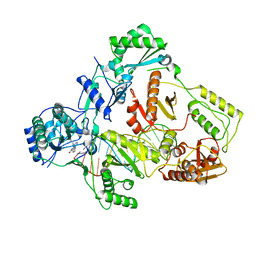 | | Cryo-EM structure of NNRTI resistant M184I/E138K mutant HIV-1 reverse transcriptase with a DNA aptamer in complex with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, DNA (38-MER), Reverse transcriptase/ribonuclease H, ... | | Authors: | Singh, A.K, Das, K. | | Deposit date: | 2022-02-26 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structures of wild-type and E138K/M184I mutant HIV-1 RT/DNA complexed with inhibitors doravirine and rilpivirine.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z2H
 
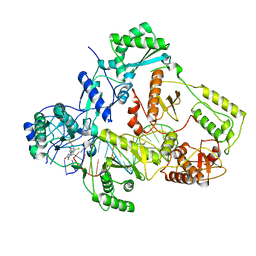 | | Cryo-EM structure of NNRTI resistant M184I/E138K mutant HIV-1 reverse transcriptase with a DNA aptamer in complex with doravirine | | Descriptor: | 3-chloro-5-({1-[(4-methyl-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl)methyl]-2-oxo-4-(trifluoromethyl)-1,2-dihydropyridin-3-yl}oxy)benzonitrile, DNA (38-MER), Reverse transcriptase/ribonuclease H, ... | | Authors: | Singh, A.K, Das, K. | | Deposit date: | 2022-02-27 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Cryo-EM structures of wild-type and E138K/M184I mutant HIV-1 RT/DNA complexed with inhibitors doravirine and rilpivirine.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z2G
 
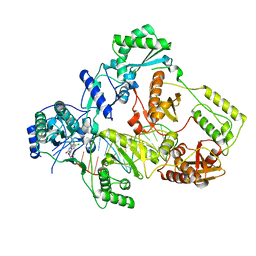 | | Cryo-EM structure of HIV-1 reverse transcriptase with a DNA aptamer in complex with doravirine | | Descriptor: | 3-chloro-5-({1-[(4-methyl-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl)methyl]-2-oxo-4-(trifluoromethyl)-1,2-dihydropyridin-3-yl}oxy)benzonitrile, DNA (38-MER), Reverse transcriptase/ribonuclease H | | Authors: | Singh, A.K, Das, K. | | Deposit date: | 2022-02-26 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Cryo-EM structures of wild-type and E138K/M184I mutant HIV-1 RT/DNA complexed with inhibitors doravirine and rilpivirine.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z70
 
 | |
6UJZ
 
 | | HIV-1 wild-type reverse transcriptase-DNA complex with (+)-FTC-TP | | Descriptor: | MAGNESIUM ION, SULFATE ION, [[(2~{S},5~{R})-5-(4-azanyl-5-fluoranyl-2-oxidanylidene-pyrimidin-1-yl)-1,3-oxathiolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2019-10-03 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55642557 Å) | | Cite: | Elucidating molecular interactions ofL-nucleotides with HIV-1 reverse transcriptase and mechanism of M184V-caused drug resistance.
Commun Biol, 2, 2019
|
|
4URE
 
 | | Molecular Genetic and Crystal Structural Analysis of 1-(4- Hydroxyphenyl)-Ethanol Dehydrogenase from Aromatoleum aromaticum EbN1 | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ACETATE ION, CYCLOHEXANOL DEHYDROGENASE, ... | | Authors: | Buesing, I, Hoeffken, H.W, Breuer, M, Woehlbrand, L, Hauer, B, Rabus, R. | | Deposit date: | 2014-06-28 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Genetic and Crystal Structural Analysis of 1-(4-Hydroxyphenyl)-Ethanol Dehydrogenase from 'Aromatoleum Aromaticum' Ebn1.
J.Mol.Microbiol.Biotechnol., 25, 2015
|
|
8JZN
 
 | | Structure of a fungal 1,3-beta-glucan synthase | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,3-beta-glucan synthase component FKS1, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, C, You, Z, Chen, D, Hang, J, Wang, Z, Meng, J, Wang, L, Zhao, P, Qiao, J, Yun, C, Bai, L. | | Deposit date: | 2023-07-05 | | Release date: | 2023-10-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structure of a fungal 1,3-beta-glucan synthase.
Sci Adv, 9, 2023
|
|
4WE1
 
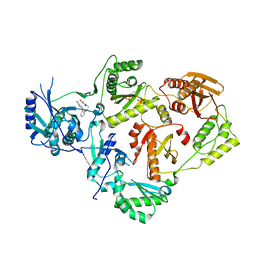 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-2-naphthonitrile (JLJ600) | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}naphthalene-2-carbonitrile, Gag-Pol polyprotein, MAGNESIUM ION, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2014-09-09 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Picomolar Inhibitors of HIV-1 Reverse Transcriptase: Design and Crystallography of Naphthyl Phenyl Ethers.
Acs Med.Chem.Lett., 5, 2014
|
|
4URF
 
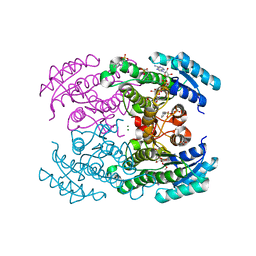 | | Molecular Genetic and Crystal Structural Analysis of 1-(4- Hydroxyphenyl)-Ethanol Dehydrogenase from Aromatoleum aromaticum EbN1 | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ACETATE ION, BICARBONATE ION, ... | | Authors: | Buesing, I, Hoeffken, H.W, Breuer, M, Woehlbrand, L, Hauer, B, Rabus, R. | | Deposit date: | 2014-06-28 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular Genetic and Crystal Structural Analysis of 1-(4-Hydroxyphenyl)-Ethanol Dehydrogenase from 'Aromatoleum Aromaticum' Ebn1.
J.Mol.Microbiol.Biotechnol., 25, 2015
|
|
1FO2
 
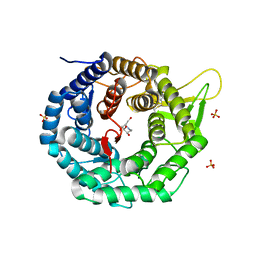 | | CRYSTAL STRUCTURE OF HUMAN CLASS I ALPHA1,2-MANNOSIDASE IN COMPLEX WITH 1-DEOXYMANNOJIRIMYCIN | | Descriptor: | 1-DEOXYMANNOJIRIMYCIN, ALPHA1,2-MANNOSIDASE, CALCIUM ION, ... | | Authors: | Vallee, F, Karaveg, K, Moremen, K.W, Herscovics, A, Howell, P.L. | | Deposit date: | 2000-08-24 | | Release date: | 2001-01-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis for catalysis and inhibition of N-glycan processing class I alpha 1,2-mannosidases.
J.Biol.Chem., 275, 2000
|
|
7P4D
 
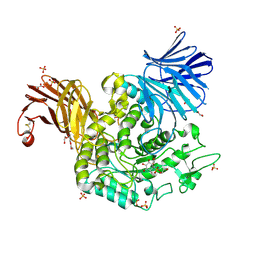 | | Crystal Structure of Agd31B, alpha-transglucosylase in Glycoside Hydrolase Family 31, in complex with covalent Cyclophellitol Sulfamidate probe KK130 | | Descriptor: | 1,2-ETHANEDIOL, OXALATE ION, Oligosaccharide 4-alpha-D-glucosyltransferase, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1,6- epi-Cyclophellitol Cyclosulfamidate Is a Bona Fide Lysosomal alpha-Glucosidase Stabilizer for the Treatment of Pompe Disease.
J.Am.Chem.Soc., 144, 2022
|
|
1G0I
 
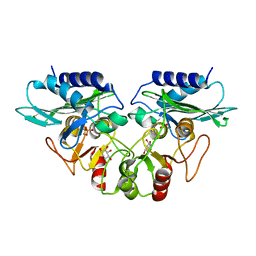 | | CRYSTAL STRUCTURE OF MJ0109 GENE PRODUCT INOSITOL MONOPHOSPHATASE-FRUCTOSE 1,6 BISPHOSPHATASE | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, INOSITOL MONOPHOSPHATASE, MANGANESE (II) ION, ... | | Authors: | Johnson, K.A, Chen, L, Yang, H, Roberts, M.F, Stec, B. | | Deposit date: | 2000-10-06 | | Release date: | 2001-03-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and catalytic mechanism of the MJ0109 gene product: a bifunctional enzyme with inositol monophosphatase and fructose 1,6-bisphosphatase activities.
Biochemistry, 40, 2001
|
|
8FNL
 
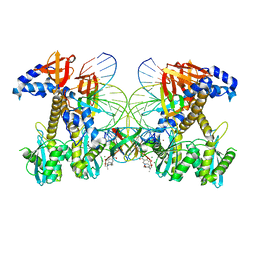 | | Structure of E138K/Q148K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNN
 
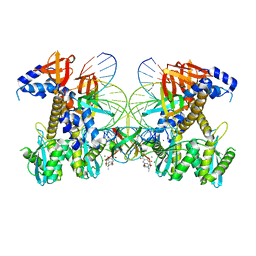 | | Structure of E138K/G140A/Q148K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNJ
 
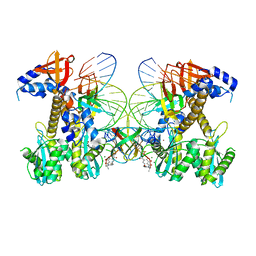 | | Structure of E138K/G140A HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNM
 
 | | Structure of G140A/Q148K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FND
 
 | | Structure of E138K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNQ
 
 | | Structure of E138K/G140A/Q148K HIV-1 intasome with 4d bound | | Descriptor: | 4-amino-N-[(2,4-difluorophenyl)methyl]-1-hydroxy-6-(6-hydroxyhexyl)-2-oxo-1,2-dihydro-1,8-naphthyridine-3-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNH
 
 | | Structure of Q148K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNP
 
 | | Structure of E138K/G140S/Q148H HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
