9EN2
 
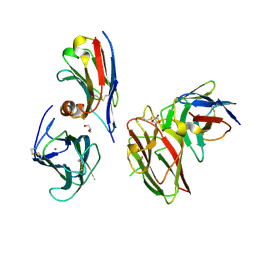 | | Crystal structure of the metalloproteinase enhancer PCPE-1 complexed with nanobodies VHH-H4 and VHH-I5 | | Descriptor: | CALCIUM ION, GLYCEROL, Procollagen C-endopeptidase enhancer 1, ... | | Authors: | Lagoutte, P, Gueguen-Chaignon, V, Bourhis, J.-M, Vadon-Le Goff, S. | | Deposit date: | 2024-03-12 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mono- and Bi-specific Nanobodies Targeting the CUB Domains of PCPE-1 Reduce the Proteolytic Processing of Fibrillar Procollagens.
J.Mol.Biol., 436, 2024
|
|
9EMV
 
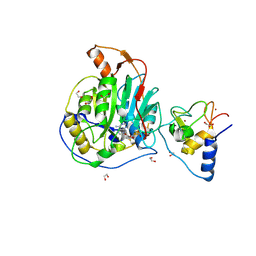 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with Sangivamycin and m7GpppA (Cap0-analog)/m7GpppAm (Cap1-analog) | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Scheer, T.E.S. | | Deposit date: | 2024-03-11 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
To Be Published
|
|
9EML
 
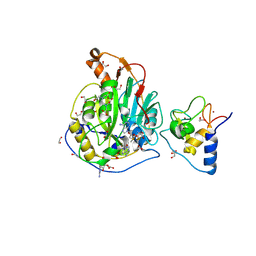 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and m7GpppA (Cap0-analog)/m7GpppAm (Cap1-analog) | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-03-08 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
To Be Published
|
|
9EMJ
 
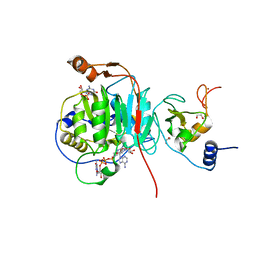 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with Toyocamycin and m7GpppA (Cap0-analog) | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 4-amino-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-03-08 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
To Be Published
|
|
9EME
 
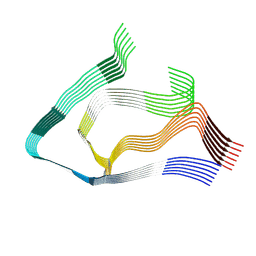 | | AL amyloid fibril from the FOR103 light chain | | Descriptor: | lambda 3 immunoglobulin light chain fragment, residues 2-116 | | Authors: | Pfeiffer, P.B, Karimi-Farsijani, S, Kupfer, N, Schmidt, M, Faendrich, M. | | Deposit date: | 2024-03-08 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Light chain mutations contribute to defining the fibril morphology in systemic AL amyloidosis.
Nat Commun, 15, 2024
|
|
9EMC
 
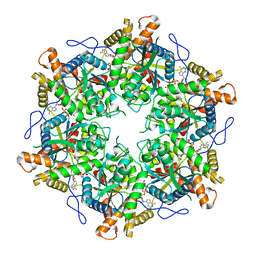 | |
9EMB
 
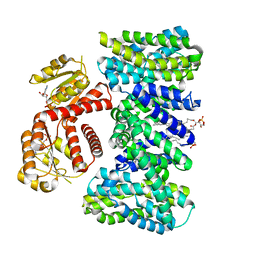 | | Structure of KefC Asp156Asn variant | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, ADENOSINE MONOPHOSPHATE, Glutathione-regulated potassium-efflux system protein KefC | | Authors: | Gulati, A, Kokane, S, Drew, D. | | Deposit date: | 2024-03-07 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structure of KefC Asp156Asn variant
To Be Published
|
|
9EMA
 
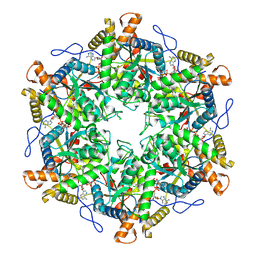 | | RUVBL1/2 in complex with ATP and CB-6644 inhibitor | | Descriptor: | 5-chloranyl-2-ethoxy-4-fluoranyl-~{N}-[4-[[3-(methoxymethyl)-1-oxidanylidene-6,7-dihydro-5~{H}-pyrazolo[1,2-a][1,2]benzodiazepin-2-yl]amino]-2,2-dimethyl-4-oxidanylidene-butyl]benzamide, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lopez-Perrote, A, Llorca, O, Garcia-Martin, C. | | Deposit date: | 2024-03-07 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Mechanism of allosteric inhibition of RUVBL1-RUVBL2 by the small-molecule CB-6644
Cell Rep Phys Sci, 2024
|
|
9EM1
 
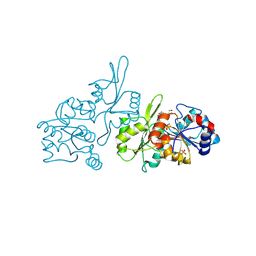 | | Human pyridoxal phosphatase in complex with 7,8-dihydroxyflavone and phosphate | | Descriptor: | 7,8-bis(oxidanyl)-2-phenyl-chromen-4-one, Chronophin, GLYCEROL, ... | | Authors: | Brenner, M, Gohla, A, Schindelin, H. | | Deposit date: | 2024-03-07 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 7,8-Dihydroxyflavone is a direct inhibitor of human and murine pyridoxal phosphatase.
Elife, 13, 2024
|
|
9DNA
 
 | |
9CKV
 
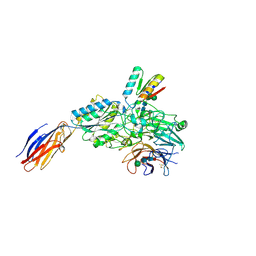 | | Cryo-EM structure of alpha5beta1 integrin in complex with NeoNectin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Werther, R, Nguyen, A, Estrada Alamo, K.A, Wang, X, Campbell, M.G. | | Deposit date: | 2024-07-09 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | De Novo Design of Integrin alpha5beta1 Modulating Proteins for Regenerative Medicine
To Be Published
|
|
9CK0
 
 | |
9CJA
 
 | |
9CGT
 
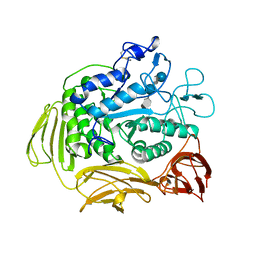 | | STRUCTURE OF CYCLODEXTRIN GLYCOSYLTRANSFERASE COMPLEXED WITH A THIO-MALTOPENTAOSE | | Descriptor: | CALCIUM ION, PROTEIN (CYCLODEXTRIN-GLYCOSYLTRANSFERASE), alpha-D-glucopyranose-(1-4)-4-thio-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4-thio-alpha-D-glucopyranose-(1-4)-1-thio-alpha-D-glucopyranose | | Authors: | Schmidt, A.K, Schulz, G.E. | | Deposit date: | 1998-09-27 | | Release date: | 1998-10-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate binding to a cyclodextrin glycosyltransferase and mutations increasing the gamma-cyclodextrin production.
Eur.J.Biochem., 255, 1998
|
|
9CGM
 
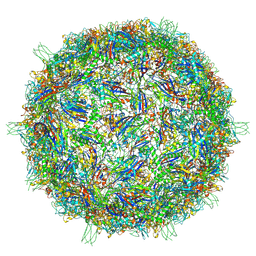 | | The Structure of Spiroplasma Virus 4 | | Descriptor: | Capsid protein VP1, DNA binding protein ORF8 | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2024-06-30 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | The Structure of Spiroplasma Virus 4: Exploring the Capsid Diversity of the Microviridae
Viruses, 2024
|
|
9CEQ
 
 | |
9CE9
 
 | |
9CBE
 
 | |
9CA2
 
 | |
9CA1
 
 | | Human TOP3B-TDRD3 core complex in DNA religation state | | Descriptor: | DNA (5'-D(*AP*TP*T)-3'), DNA (5'-D(P*TP*AP*CP*TP*AP*AP*A)-3'), DNA topoisomerase 3-beta-1, ... | | Authors: | Yang, X, Chen, X, Yang, W, Pommier, Y. | | Deposit date: | 2024-06-16 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural Insights into human Topoisomerase 3-beta DNA and RNA catalytic cycles and topo-gate dynamics
To Be Published
|
|
9CA0
 
 | | Human TOP3B-TDRD3 core complex in DNA pre-cleavage state | | Descriptor: | DNA (5'-D(*AP*TP*T)-3'), DNA (5'-D(P*TP*AP*CP*TP*AP*AP*A)-3'), DNA topoisomerase 3-beta-1, ... | | Authors: | Yang, X, Chen, X, Yang, W, Pommier, Y. | | Deposit date: | 2024-06-16 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural Insights into human Topoisomerase 3-beta DNA and RNA catalytic cycles and topo-gate dynamics
To Be Published
|
|
9C9Y
 
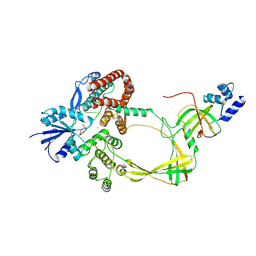 | | Human TOP3B-TDRD3 core complex in DNA pre-cleavage state | | Descriptor: | DNA (5'-D(P*AP*CP*TP*AP*AP*AP*AP*T)-3'), DNA topoisomerase 3-beta-1, MANGANESE (II) ION, ... | | Authors: | Yang, X, Chen, X, Yang, W, Pommier, Y. | | Deposit date: | 2024-06-16 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural Insights into human Topoisomerase 3-beta DNA and RNA catalytic cycles and topo-gate dynamics
To Be Published
|
|
9C8S
 
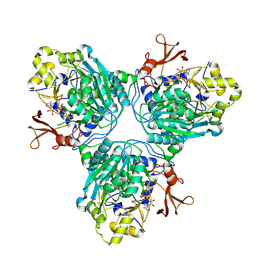 | |
9C8R
 
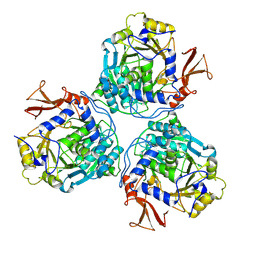 | |
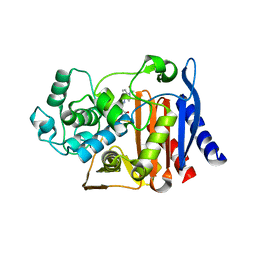
9C8J
 
 | |
