6ZNK
 
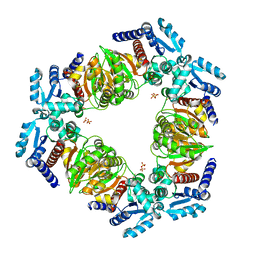 | | MaeB PTA domain N718D mutant | | Descriptor: | Malate dehydrogenase, SULFATE ION | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2020-07-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.039 Å) | | Cite: | A rotary mechanism for allostery in bacterial hybrid malic enzymes.
Nat Commun, 12, 2021
|
|
2LRS
 
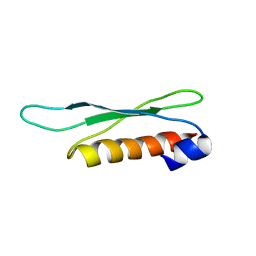 | | The second dsRBD domain from A. thaliana DICER-LIKE 1 | | Descriptor: | Endoribonuclease Dicer homolog 1 | | Authors: | Burdisso, P, Suarez, I, Bersch, B, Bologna, N, Palatnik, J, Boisbouvier, J, Rasia, R. | | Deposit date: | 2012-04-12 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Second Double-Stranded RNA Binding Domain of Dicer-like Ribonuclease 1: Structural and Biochemical Characterization.
Biochemistry, 51, 2012
|
|
7SL6
 
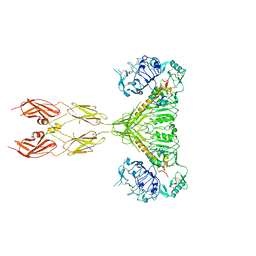 | |
5JW1
 
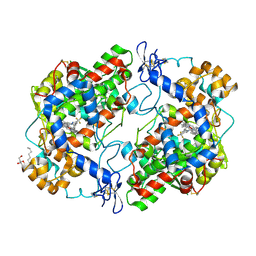 | | Crystal structure of Celecoxib bound to S121P murine COX-2 mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[5-(4-METHYLPHENYL)-3-(TRIFLUOROMETHYL)-1H-PYRAZOL-1-YL]BENZENESULFONAMIDE, ... | | Authors: | Malkowski, M.G, Orlando, B.J. | | Deposit date: | 2016-05-11 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.822 Å) | | Cite: | Fatty Acid Binding to the Allosteric Subunit of Cyclooxygenase-2 Relieves a Tonic Inhibition of the Catalytic Subunit.
J.Biol.Chem., 291, 2016
|
|
7A8B
 
 | |
7SL7
 
 | |
2YYG
 
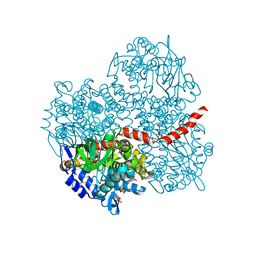 | | Crystal structure of the oxygenase component (HpaB) of 4-hydroxyphenylacetate 3-monooxygenase | | Descriptor: | 4-hydroxyphenylacetate-3-hydroxylase, SULFATE ION | | Authors: | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-04-30 | | Release date: | 2007-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
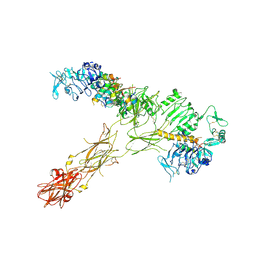
7SL2
 
 | |
7S06
 
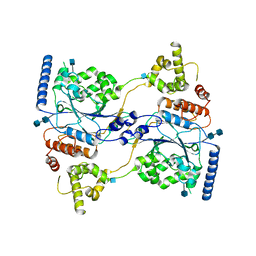 | | Cryo-EM structure of human GlcNAc-1-phosphotransferase A2B2 subcomplex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetylglucosamine-1-phosphotransferase subunits alpha/beta | | Authors: | Li, H, Li, H. | | Deposit date: | 2021-08-30 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the human GlcNAc-1-phosphotransferase alpha beta subunits reveals regulatory mechanism for lysosomal enzyme glycan phosphorylation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
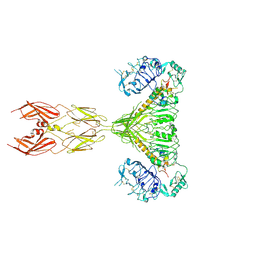
7SL3
 
 | |
8OYG
 
 | |
2G82
 
 | |
5JXI
 
 | | Structure of the unliganded form of the proprotein convertase furin in presence of EDTA. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Furin, ... | | Authors: | Dahms, S.O, Arciniega, M, Steinmetzer, T, Huber, R, Than, M.E. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2YZM
 
 | |
7SL4
 
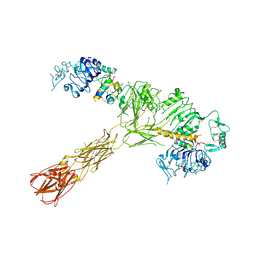 | |
2LGY
 
 | | Ubiquitin-like domain from HOIL-1 | | Descriptor: | RanBP-type and C3HC4-type zinc finger-containing protein 1 | | Authors: | Beasley, S.A, Shaw, G.S. | | Deposit date: | 2011-08-03 | | Release date: | 2012-06-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the E3 ligase HOIL-1 Ubl domain.
Protein Sci., 21, 2012
|
|
7SFE
 
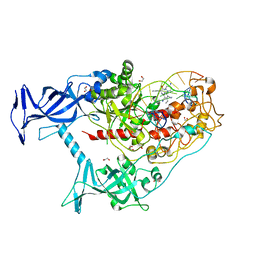 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Inhibitor GSK3830334A | | Descriptor: | (2R)-2-{[6-(4-aminopiperidin-1-yl)-3,5-dicyano-4-ethylpyridin-2-yl]amino}-2-phenylacetamide, 1,2-ETHANEDIOL, DNA (12-MER), ... | | Authors: | Horton, J.R, Pathuri, S, Cheng, X. | | Deposit date: | 2021-10-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural characterization of dicyanopyridine containing DNMT1-selective, non-nucleoside inhibitors.
Structure, 30, 2022
|
|
7S4M
 
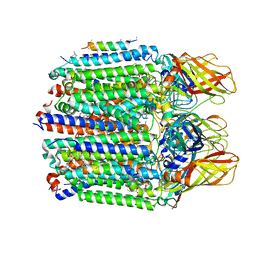 | |
2LLS
 
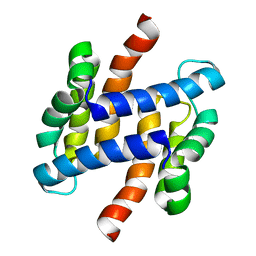 | | solution structure of human apo-S100A1 C85M | | Descriptor: | Protein S100-A1 | | Authors: | Budzinska, M, Jaremko, L, Jaremko, M, Zdanowski, K, Zhukov, I, Bierzynski, A, Ejchart, A. | | Deposit date: | 2011-11-17 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Chemical Shift Assignments and solution structure of human apo-S100A1 C85M mutant
To be Published
|
|
7SFG
 
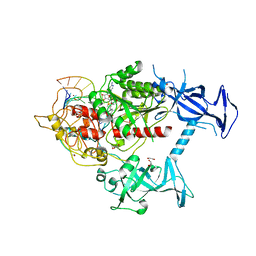 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Cofactor SAM | | Descriptor: | 1,2-ETHANEDIOL, DNA (cytosine-5)-methyltransferase 1, DNA Strand 1, ... | | Authors: | Horton, J.R, Pathuri, S, Cheng, X. | | Deposit date: | 2021-10-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural characterization of dicyanopyridine containing DNMT1-selective, non-nucleoside inhibitors.
Structure, 30, 2022
|
|
2LO3
 
 | | Solution structure of Sgf73(59-102) zinc finger domain | | Descriptor: | SAGA-associated factor 73, ZINC ION | | Authors: | Gao, X, Koehler, C, Bonnet, J, Devys, D, Kieffer, B. | | Deposit date: | 2012-01-10 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into the role of SGF11 and SGF73 for the interaction between SAGA and nucleosomes
To be Published
|
|
6ZNG
 
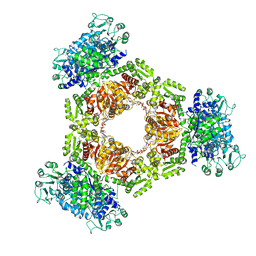 | | MaeB full-length acetyl-CoA bound state | | Descriptor: | ACETYL COENZYME *A, NADP-dependent malate dehydrogenase,Malate dehydrogenase | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2020-07-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A rotary mechanism for allostery in bacterial hybrid malic enzymes.
Nat Commun, 12, 2021
|
|
7DDQ
 
 | | Structure of RC-LH1-PufX from Rhodobacter veldkampii | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Antenna pigment protein alpha chain, Antenna pigment protein beta chain, ... | | Authors: | Bracun, L, Yamagata, A, Shirouzu, M, Liu, L.N. | | Deposit date: | 2020-10-29 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of the photosynthetic RC-LH1-PufX supercomplex at 2.8-angstrom resolution.
Sci Adv, 7, 2021
|
|
2LOJ
 
 | | Solution NMR structure of TSTM1273 from Salmonella typhimurium LT2, NESG target STT322, CSGID target IDP01027 and OCSP target TSTM1273 | | Descriptor: | Putative cytoplasmic protein | | Authors: | Wu, B, Yee, A, Houliston, S, Garcia, M, Savchenko, A, Arrowsmith, C.H, Anderson, W.F, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of TSTM1273 from Salmonella typhimurium LT2, NESG target STT322, CSGID target IDP01027 and OCSP target TSTM1273
To be Published
|
|
7DL8
 
 | |
