7P13
 
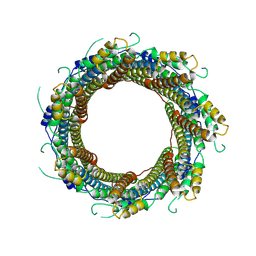 | | 2.29 A Mycobacterium tuberculosis EspB. | | Descriptor: | ESX-1 secretion-associated protein EspB | | Authors: | Gijsbers, A, Zhang, Y, Vinciauskaite, V, Siroy, A, Gao, Y, Tria, G, Mathew, A, Sanchez-Puig, N, Lopez-Iglesias, C, Peters, P.J, Ravelli, R.B.G. | | Deposit date: | 2021-07-01 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Priming mycobacterial ESX-secreted protein B to form a channel-like structure.
Curr Res Struct Biol, 3, 2021
|
|
5KKD
 
 | |
6IM8
 
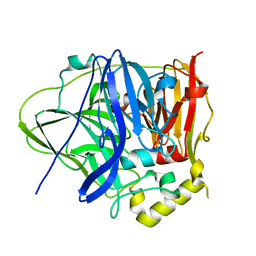 | | CueO-PM2 multicopper oxidase | | Descriptor: | Blue copper oxidase CueO,PM2 peptide,Blue copper oxidase CueO | | Authors: | Wongsantichon, J, Robinson, R, Ghadessy, F. | | Deposit date: | 2018-10-22 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Development and structural characterization of an engineered multi-copper oxidase reporter of protein-protein interactions.
J.Biol.Chem., 294, 2019
|
|
4ZH2
 
 | |
5GXU
 
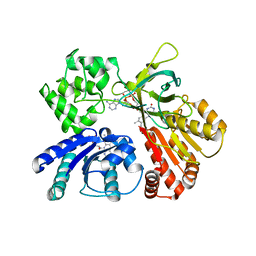 | | Cystal structure of Arabidopsis ATR2 | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH--cytochrome P450 reductase 2 | | Authors: | Niu, G, Liu, L. | | Deposit date: | 2016-09-20 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Arabidopsis thaliana NADPH-cytochrome P450 reductase 2 (ATR2) provides insight into its function
FEBS J., 284, 2017
|
|
5XNL
 
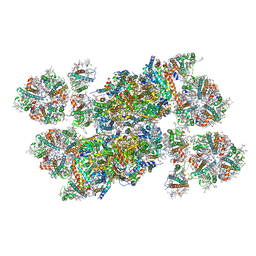 | | Structure of stacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
6IBD
 
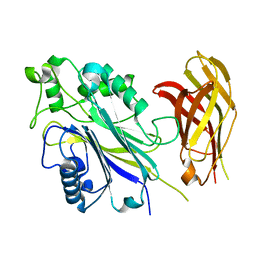 | | The Phosphatase and C2 domains of Human SHIP1 | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Williams, E.P, Fernandez-Cid, A, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2018-11-29 | | Release date: | 2019-01-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
6RZW
 
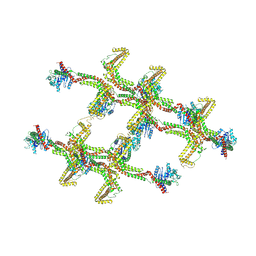 | | Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes in the GTPgammaS bound state | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (18.799999 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
4ZOK
 
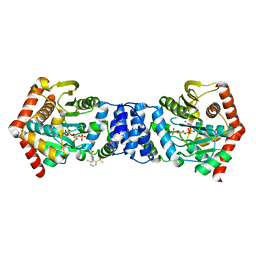 | |
6URE
 
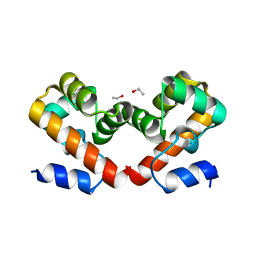 | | Barrier-to-autointegration factor Aqueous: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-23 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5KSR
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa - XFSurE-TB (Tetramer Bigger). | | Descriptor: | 5'-nucleotidase SurE, CHLORIDE ION, IODIDE ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, De Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
6URK
 
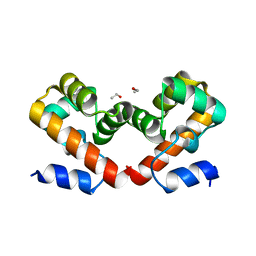 | | Barrier-to-autointegration factor soaked in Glycerol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-23 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6IBH
 
 | | Copper binding protein from Laetisaria arvalis (LaX325) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity CAZyme, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Tandrup, T, Labourel, A, Haon, M, Berrin, J.-G, Lo Leggio, L. | | Deposit date: | 2018-11-30 | | Release date: | 2019-11-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A fungal family of lytic polysaccharide monooxygenase-like copper proteins.
Nat.Chem.Biol., 16, 2020
|
|
4R7Y
 
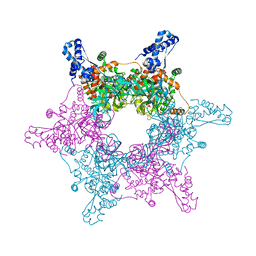 | | Crystal structure of an active MCM hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Miller, J.M, Arachea, B.T, Epling, L.B, Enemark, E.J. | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Analysis of the crystal structure of an active MCM hexamer.
Elife, 3, 2014
|
|
7OWG
 
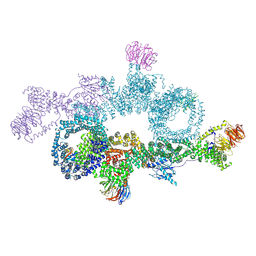 | | human DEPTOR in a complex with mutant human mTORC1 A1459P | | Descriptor: | DEP domain-containing mTOR-interacting protein, Regulatory-associated protein of mTOR, Serine/threonine-protein kinase mTOR, ... | | Authors: | Heimhalt, M, Berndt, A, Wagstaff, J, Anandapadamanaban, M, Perisic, O, Maslen, S, McLaughlin, S, Yu, W.-H, Masson, G.R, Boland, A, Ni, X, Yamashita, K, Murshudov, G.N, Skehel, M, Freund, S.M, Williams, R.L. | | Deposit date: | 2021-06-18 | | Release date: | 2021-09-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Bipartite binding and partial inhibition links DEPTOR and mTOR in a mutually antagonistic embrace.
Elife, 10, 2021
|
|
6S0O
 
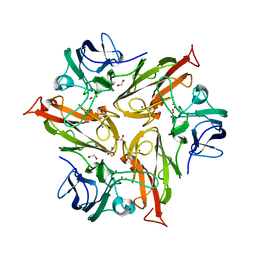 | | Crystal Structure of Two-Domain Laccase from Streptomyces griseoflavus produced at 0.25 mM copper sulfate in growth medium | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Kolyadenko, I.A. | | Deposit date: | 2019-06-17 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Investigations of Accessibility of T2/T3 Copper Center of Two-Domain Laccase fromStreptomyces griseoflavusAc-993.
Int J Mol Sci, 20, 2019
|
|
6UPZ
 
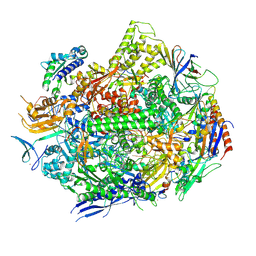 | | RNA polymerase II elongation complex with 5-guanidinohydantoin lesion in state 3 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2019-10-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair-pi and CH-pi interactions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3PXS
 
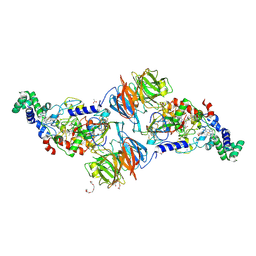 | |
5U8L
 
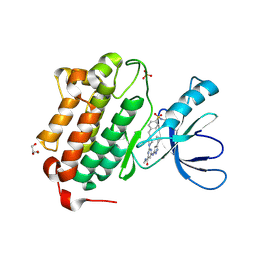 | | Crystal structure of EGFR kinase domain in complex with a sulfonyl fluoride probe XO44 | | Descriptor: | 4-[(4-{4-[(3-cyclopropyl-1H-pyrazol-5-yl)amino]-6-[(prop-2-yn-1-yl)carbamoyl]pyrimidin-2-yl}piperazin-1-yl)methyl]benzene-1-sulfonyl fluoride, Epidermal growth factor receptor, GLYCEROL, ... | | Authors: | Gajiwala, K.S, Ferre, R. | | Deposit date: | 2016-12-14 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Broad-Spectrum Kinase Profiling in Live Cells with Lysine-Targeted Sulfonyl Fluoride Probes.
J. Am. Chem. Soc., 139, 2017
|
|
3LK2
 
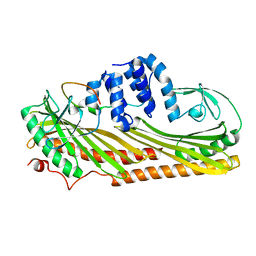 | | Crystal structure of CapZ bound to the uncapping motif from CARMIL | | Descriptor: | F-actin-capping protein subunit alpha-1, F-actin-capping protein subunit beta isoforms 1 and 2, Leucine-rich repeat-containing protein 16A | | Authors: | Hernandez-Valladares, M, Kim, T, Kannan, B, Tung, A, Aguda, A.H, Larsson, M, Cooper, J.A, Robinson, R.C. | | Deposit date: | 2010-01-27 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of a capping protein interaction motif defines a family of actin filament regulators.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7OZM
 
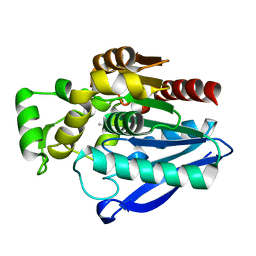 | | Crystal Structure of mtbMGL K74A (Closed Cap Conformation) | | Descriptor: | ISOPROPYL ALCOHOL, Monoacylglycerol lipase | | Authors: | Grininger, C, Aschauer, P, Pavkov-Keller, T, Oberer, M. | | Deposit date: | 2021-06-28 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Changes in the Cap of Rv0183/mtbMGL Modulate the Shape of the Binding Pocket.
Biomolecules, 11, 2021
|
|
7P0Y
 
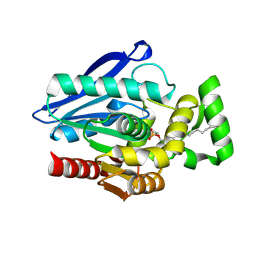 | | Crystal Structure of mtbMGL K74A (Substrate Analog Complex) | | Descriptor: | 1-[butyl(fluoranyl)phosphoryl]oxyhexadecane, Monoacylglycerol lipase | | Authors: | Grininger, C, Aschauer, P, Pavkov-Keller, T, Oberer, M. | | Deposit date: | 2021-06-30 | | Release date: | 2021-09-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Changes in the Cap of Rv0183/mtbMGL Modulate the Shape of the Binding Pocket.
Biomolecules, 11, 2021
|
|
2M43
 
 | | DNA containing a cluster of 8-oxo-guanine and abasic site lesion: alpha anomer (AP6, 8OG 14) | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*CP*(ORP)P*CP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*(8OG)P*TP*GP*GP*GP*AP*GP*CP*G)-3') | | Authors: | Zalesak, J, Jourdan, M, Constant, J, Lourdin, M. | | Deposit date: | 2013-01-29 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of DNA duplexes containing a cluster of mutagenic 8-oxoguanine and abasic site lesions.
J.Mol.Biol., 426, 2014
|
|
6USD
 
 | | Barrier-to-autointegration factor soaked in ethanol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-25 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
2M6Z
 
 | |
