5OXI
 
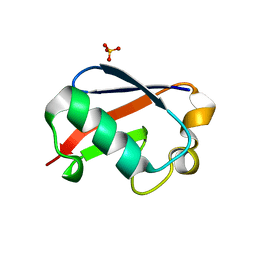 | | C-terminally retracted ubiquitin L67S mutant | | Descriptor: | SULFATE ION, Ubiquitin L67S mutant | | Authors: | Gladkova, C.G, Schubert, A.F, Wagstaff, J.L, Pruneda, J.N, Freund, S.M.V, Komander, D. | | Deposit date: | 2017-09-06 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | An invisible ubiquitin conformation is required for efficient phosphorylation by PINK1.
EMBO J., 36, 2017
|
|
6TUH
 
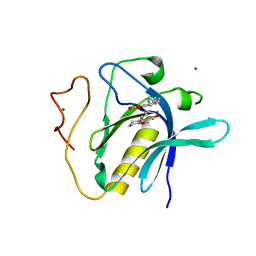 | | PH domain of Bruton's tyrosine kinase bound to compound 1 | | Descriptor: | 4,5,6,7-tetrahydro-1-benzofuran-3-carboxylic acid, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of 1-methylindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C
To Be Published
|
|
6TPK
 
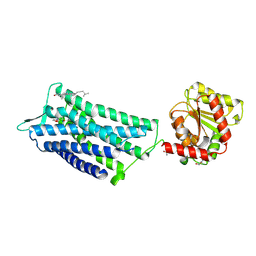 | | Crystal structure of the human oxytocin receptor | | Descriptor: | (3~{R},6~{R})-6-[(2~{S})-butan-2-yl]-3-(2,3-dihydro-1~{H}-inden-2-yl)-1-[(1~{R})-1-(2-methyl-1,3-oxazol-4-yl)-2-morpholin-4-yl-2-oxidanylidene-ethyl]piperazine-2,5-dione, CHOLESTEROL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Waltenspuehl, Y, Schoeppe, J, Ehrenmann, J, Kummer, L, Plueckthun, A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-08-05 | | Last modified: | 2020-09-02 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the human oxytocin receptor.
Sci Adv, 6, 2020
|
|
5OYD
 
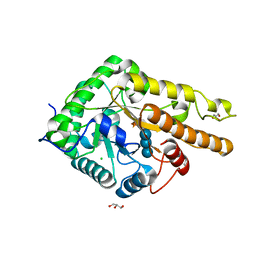 | | GH5 endo-xyloglucanase from Cellvibrio japonicus | | Descriptor: | CHLORIDE ION, Cellulase, putative, ... | | Authors: | Attia, M, Nelson, C.E, Offen, W.A, Jain, N, Gardner, J.G, Davies, G.J, Brumer, H. | | Deposit date: | 2017-09-08 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | In vitro and in vivo characterization of threeCellvibrio japonicusglycoside hydrolase family 5 members reveals potent xyloglucan backbone-cleaving functions.
Biotechnol Biofuels, 11, 2018
|
|
6TRH
 
 | |
6TT4
 
 | |
6TTK
 
 | | Crystal structure of the kelch domain of human KLHL12 in complex with DVL1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DVL1, ... | | Authors: | Chen, Z, Williams, E, Pike, A.C.W, Strain-Damerell, C, Wang, D, Chalk, R, Burgess-Brown, N, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-12-27 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | Identification of a PGXPP degron motif in dishevelled and structural basis for its binding to the E3 ligase KLHL12.
Open Biology, 10, 2020
|
|
6TUA
 
 | | The RYK Pseudokinase Domain | | Descriptor: | SULFATE ION, Tyrosine-protein kinase RYK | | Authors: | Mathea, S, Chatterjee, D, Preuss, F, Shin, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2020-01-04 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural Insights into Pseudokinase Domains of Receptor Tyrosine Kinases.
Mol.Cell, 79, 2020
|
|
5OTI
 
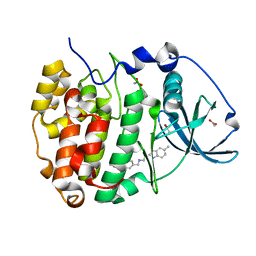 | | The crystal structure of CK2alpha in complex with compound 27 | | Descriptor: | ACETATE ION, Casein kinase II subunit alpha, ~{N}-[[3-chloranyl-4-(2-ethylphenyl)phenyl]methyl]-2-(5-methyl-1~{H}-benzimidazol-2-yl)ethanamine | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-08-22 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
5OTN
 
 | | Crystal structure of zebrafish MTH1 in complex with O6-methyl-dGMP | | Descriptor: | 1,2-ETHANEDIOL, 6-O-METHYL GUANOSINE-5'-MONOPHOSPHATE, CALCIUM ION, ... | | Authors: | Gustafsson, R, Henriksson, L, Jemth, A.-S, Brautigam, L, Carreras Puigvert, J, Homan, E, Warpman Berglund, U, Helleday, T, Stenmark, P. | | Deposit date: | 2017-08-22 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | MutT homologue 1 (MTH1) catalyzes the hydrolysis of mutagenic O6-methyl-dGTP.
Nucleic Acids Res., 46, 2018
|
|
6TUM
 
 | |
6T86
 
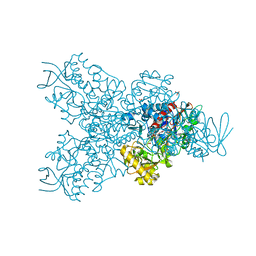 | | Urocanate reductase in complex with FAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Venskutonyte, R, Lindkvist-Petersson, K. | | Deposit date: | 2019-10-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural characterization of the microbial enzyme urocanate reductase mediating imidazole propionate production.
Nat Commun, 12, 2021
|
|
6T87
 
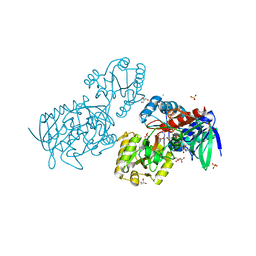 | | Urocanate reductase in complex with urocanate | | Descriptor: | (2E)-3-(1H-IMIDAZOL-4-YL)ACRYLIC ACID, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Venskutonyte, R, Lindkvist-Petersson, K. | | Deposit date: | 2019-10-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural characterization of the microbial enzyme urocanate reductase mediating imidazole propionate production.
Nat Commun, 12, 2021
|
|
5OTY
 
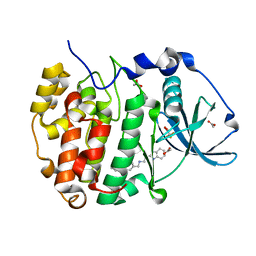 | | The crystal structure of CK2alpha in complex with CAM4712 | | Descriptor: | 2-(1~{H}-benzimidazol-2-yl)-~{N}-[[3,5-bis(chloranyl)-4-(2-ethylphenyl)phenyl]methyl]ethanamine, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-08-22 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
5OU9
 
 | |
6T88
 
 | |
6T85
 
 | | Urocanate reductase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Venskutonyte, R, Lindkvist-Petersson, K. | | Deposit date: | 2019-10-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural characterization of the microbial enzyme urocanate reductase mediating imidazole propionate production.
Nat Commun, 12, 2021
|
|
6T1V
 
 | | Structure of PPARg H494Y mutant in complex with GW1929 | | Descriptor: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, Peroxisome proliferator-activated receptor gamma, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | Authors: | Rochel, N. | | Deposit date: | 2019-10-06 | | Release date: | 2021-04-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of a PPARg mutant complex
To Be Published
|
|
6T1S
 
 | | PPAR mutant | | Descriptor: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, Peroxisome proliferator-activated receptor gamma, SULFATE ION | | Authors: | Rochel, N. | | Deposit date: | 2019-10-05 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of PPARg mutant
To Be Published
|
|
6T2A
 
 | |
6TE3
 
 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Cocrystal with Fe2+, Mn2+, UDP | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chiapparino, A, De Giorgi, F, Scietti, L, Faravelli, S, Roscioli, T, Forneris, F. | | Deposit date: | 2019-11-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Regulatory Molecular 'Hot Spots' for LH/PLOD Collagen Glycosyltransferase Activity
Int J Mol Sci, 2023
|
|
6TEC
 
 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Cocrystal with Fe2+, Mn2+, UDP-Xylose | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chiapparino, A, De Giorgi, F, Scietti, L, Faravelli, S, Roscioli, T, Forneris, F. | | Deposit date: | 2019-11-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of Regulatory Molecular 'Hot Spots' for LH/PLOD Collagen Glycosyltransferase Activity
Int J Mol Sci, 2023
|
|
6TES
 
 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Cocrystal with Fe2+, Mn2+, UDP-Glucuronic Acid | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chiapparino, A, De Giorgi, F, Scietti, L, Faravelli, S, Roscioli, T, Forneris, F. | | Deposit date: | 2019-11-12 | | Release date: | 2021-05-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of Regulatory Molecular 'Hot Spots' for LH/PLOD Collagen Glycosyltransferase Activity
Int J Mol Sci, 2023
|
|
6TEU
 
 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Val80Lys mutant - Cocrystal with Fe2+, Mn2+ | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chiapparino, A, De Giorgi, F, Scietti, L, Faravelli, S, Roscioli, T, Forneris, F. | | Deposit date: | 2019-11-12 | | Release date: | 2021-05-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of Regulatory Molecular 'Hot Spots' for LH/PLOD Collagen Glycosyltransferase Activity
Int J Mol Sci, 2023
|
|
6TEZ
 
 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Val80Lys mutant - Cocrystal with Fe2+, Mn2+, UDP-Glucuronic Acid | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chiapparino, A, De Giorgi, F, Scietti, L, Faravelli, S, Roscioli, T, Forneris, F. | | Deposit date: | 2019-11-12 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of Regulatory Molecular 'Hot Spots' for LH/PLOD Collagen Glycosyltransferase Activity
Int J Mol Sci, 2023
|
|
