6TJT
 
 | | Neuropilin2-b1 domain in a complex with the C-terminal VEGFC peptide | | Descriptor: | 1,2-ETHANEDIOL, Neuropilin-2, SULFATE ION, ... | | Authors: | Eldrid, C, Yu, L, Yelland, T, Fotinou, C, Djordjevic, S. | | Deposit date: | 2019-11-26 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | NRP2-b1 domain in a complex with the C-terminal VEGFC peptide
To Be Published
|
|
5OPX
 
 | | Crystal structure of the GroEL mutant A109C in complex with GroES and ADP BeF2 | | Descriptor: | 10 kDa chaperonin, 60 kDa chaperonin, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Yan, X, Shi, Q, Bracher, A, Milicic, G, Singh, A.K, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2017-08-10 | | Release date: | 2018-01-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | GroEL Ring Separation and Exchange in the Chaperonin Reaction.
Cell, 172, 2018
|
|
6TEQ
 
 | | Crystal structure of a galactokinase from Bifidobacterium infantis in complex with 2-deoxy-2-fluoro-galactose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-deoxy-2-fluoro-alpha-D-galactopyranose, ... | | Authors: | Keenan, T, Parmeggiani, F, Fontenelle, C.Q, Malassis, J, Vendeville, J, Offen, W.A, Both, P, Huang, K, Marchesi, A, Heyam, A, Young, C, Charnock, S, Davies, G.J, Linclau, B, Flitsch, S.L, Fascione, M.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Profiling Substrate Promiscuity of Wild-Type Sugar Kinases for Multi-fluorinated Monosaccharides.
Cell Chem Biol, 27, 2020
|
|
5OS7
 
 | | The crystal structure of CK2alpha in complex with compound 4 | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-08-17 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
6TFO
 
 | | Crystal structure of as isolated three-domain copper-containing nitrite reductase from Hyphomicrobium denitrificans strain 1NES1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-14 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of substrate- and product-bound forms of a multi-domain copper nitrite reductase shed light on the role of domain tethering in protein complexes.
Iucrj, 7, 2020
|
|
5OT6
 
 | | The crystal structure of CK2alpha in complex with compound 19 | | Descriptor: | (3-chloranyl-4-phenyl-phenyl)methyl-[2-(1~{H}-pyrrol-2-yl)ethyl]azanium, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-08-21 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
5OTL
 
 | | The crystal structure of CK2alpha in complex with compound 29 | | Descriptor: | ACETATE ION, Casein kinase II subunit alpha, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-08-22 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
6TG8
 
 | | Crystal structure of the Kelch domain in complex with 11 amino acid peptide (model of the ETGE loop) | | Descriptor: | Kelch-like ECH-associated protein 1, SODIUM ION, VAL-ILE-ASN-PRO-GLU-THR-GLY-GLU-GLN-ILE-GLN | | Authors: | Kekez, I, Matic, S, Tomic, S, Matkovic-Calogovic, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Binding of dipeptidyl peptidase III to the oxidative stress cell sensor Kelch-like ECH-associated protein 1 is a two-step process.
J.Biomol.Struct.Dyn., 39, 2021
|
|
6TMT
 
 | | Crystal structure of the chaperonin gp146 from the bacteriophage EL 2 (Pseudomonas aeruginosa) in presence of ATP-BeFx, crystal form I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Putative GroEL-like chaperonine protein | | Authors: | Bracher, A, Paul, S.S, Wang, H, Wischnewski, N, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.03 Å) | | Cite: | Structure and conformational cycle of a bacteriophage-encoded chaperonin.
Plos One, 15, 2020
|
|
5OU7
 
 | |
5OUM
 
 | | The crystal structure of CK2alpha in complex with compound 21 | | Descriptor: | ACETATE ION, Casein kinase II subunit alpha, ~{N}-[(3-chloranyl-4-phenyl-phenyl)methyl]-2-(1~{H}-imidazol-2-yl)ethanamine | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-08-24 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
6TOV
 
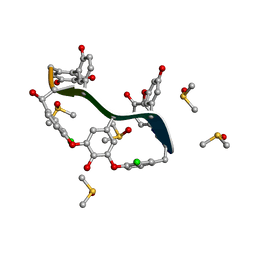 | | Crystal Structure of Teicoplanin Aglycone | | Descriptor: | DIMETHYL SULFOXIDE, Teicoplanin Aglycone | | Authors: | Belviso, B.D, Carrozzini, B, Caliandro, R, Altomare, C.D, Bolognino, I, Cellamare, S. | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-15 | | Last modified: | 2022-01-19 | | Method: | X-RAY DIFFRACTION (0.767 Å) | | Cite: | Enantiomeric Separation and Molecular Modelling of Bioactive 4-Aryl-3,4-dihydropyrimidin-2(1H)-one Ester Derivatives on Teicoplanin-Based Chiral Stationary Phase
Separations, 2022
|
|
6TID
 
 | |
6TP0
 
 | |
6TQ8
 
 | |
5OXV
 
 | | Structure of the 4_601_157 tetranucleosome (C2 form) | | Descriptor: | DNA STRAND 1 (601-based sequence model), DNA STRAND 2 (601-based sequence model), Histone H2A, ... | | Authors: | Ekundayo, B, Schalch, T. | | Deposit date: | 2017-09-07 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (6.721 Å) | | Cite: | Capturing Structural Heterogeneity in Chromatin Fibers.
J. Mol. Biol., 429, 2017
|
|
5OMT
 
 | | Endonuclease NucB | | Descriptor: | NucB | | Authors: | Basle, A, Lewis, R.J. | | Deposit date: | 2017-08-01 | | Release date: | 2017-11-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of NucB, a biofilm-degrading endonuclease.
Nucleic Acids Res., 46, 2018
|
|
6TJC
 
 | | Crystal structure of the computationally designed Cake3 protein | | Descriptor: | Cake3, GLYCEROL, PHOSPHATE ION | | Authors: | Laier, I, Mylemans, B, Voet, A.R.D, Noguchi, H. | | Deposit date: | 2019-11-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
5OMY
 
 | | HIGH-SALT STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA) IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR 4P | | Descriptor: | 4-(3-methylbut-2-enoxy)-5-propan-2-yl-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Hochscherf, J, Lindenblatt, D, Witulski, B, Birus, R, Aichele, D, Marminon, C, Bouaziz, Z, Le Borgne, M, Jose, J, Niefind, K. | | Deposit date: | 2017-08-02 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unexpected Binding Mode of a Potent Indeno[1,2-b]indole-Type Inhibitor of Protein Kinase CK2 Revealed by Complex Structures with the Catalytic Subunit CK2 alpha and Its Paralog CK2 alpha '.
Pharmaceuticals (Basel), 10, 2017
|
|
6TJG
 
 | | Crystal structure of the computationally designed Cake8 protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cake8 | | Authors: | Laier, I, Mylemans, B, Noguchi, H, Voet, A.R.D. | | Deposit date: | 2019-11-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
6TQP
 
 | | Structural insight into tanapoxvirus mediated inhibition of apoptosis | | Descriptor: | 16L protein, Bcl-2-binding component 3, isoforms 1/2, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2019-12-17 | | Release date: | 2020-06-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.84940946 Å) | | Cite: | Structural insight into tanapoxvirus-mediated inhibition of apoptosis.
Febs J., 287, 2020
|
|
6TQU
 
 | | The crystal structure of the MSP domain of human MOSPD2 in complex with the Phospho-FFAT motif of STARD3. | | Descriptor: | Motile sperm domain-containing protein 2, SULFATE ION, StAR-related lipid transfer protein 3 | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
5OOG
 
 | | Human biliverdin IX beta reductase: NADP/Phloxine B ternary complex | | Descriptor: | Flavin reductase (NADPH), GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Manso, J.A, Pereira, P.J.B. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | In silicoand crystallographic studies identify key structural features of biliverdin IX beta reductase inhibitors having nanomolar potency.
J. Biol. Chem., 293, 2018
|
|
6TLH
 
 | |
5OOU
 
 | | Designed Ankyrin Repeat Protein (DARPin) YTRL-1 selected by directed evolution against Lysozyme | | Descriptor: | DARPin YTRL-1 | | Authors: | Fischer, G, Hogan, B.J, Houlihan, G, Edmond, S, Huovinen, T.T.K, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2017-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Designed Ankyrin Repeat Protein (DARPin) YTRL-1 selected by directed evolution against Lysozyme
To be published
|
|
