5O0C
 
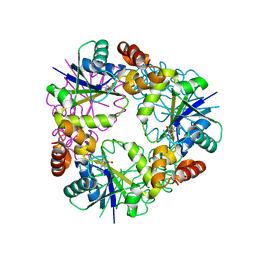 | | Crystal structure of Phosphopantetheine adenylyltransferase from Mycobacterium abcessus in complex with 3-(3-methyl-1H-indol-1-yl)propanoic acid (Fragment 3) | | Descriptor: | 3-(3-methylindol-1-yl)propanoic acid, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, Kim, S.Y, Mendes, V, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2017-05-16 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Structural Biology and the Design of New Therapeutics: From HIV and Cancer to Mycobacterial Infections: A Paper Dedicated to John Kendrew.
J. Mol. Biol., 429, 2017
|
|
5O1Y
 
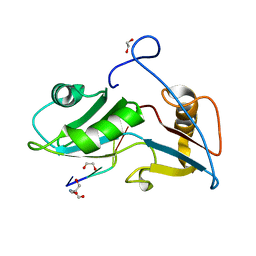 | | Structure of Nrd1 RNA binding domain in complex with RNA (GUAA) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Protein NRD1, ... | | Authors: | Franco-Echevarria, E, Perez-Canadillas, J.M, Gonzalez, B. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structure of transcription termination factor Nrd1 reveals an original mode for GUAA recognition.
Nucleic Acids Res., 45, 2017
|
|
6REH
 
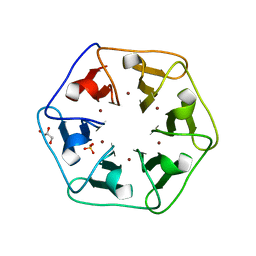 | | Crystal structure of Pizza6-S with Cu2+ | | Descriptor: | COPPER (II) ION, GLYCEROL, Pizza6-S, ... | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
6REJ
 
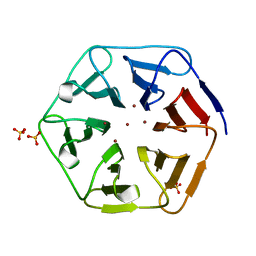 | | Crystal structure of Pizza6-SH with Zn2+ | | Descriptor: | Pizza6-SH, SULFATE ION, ZINC ION | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
6REO
 
 | | Crystal structure of 3fPizza6-SH with Sulphate ion | | Descriptor: | 3fPizza6-SH, SULFATE ION | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Artificial beta-Propeller Protein-based Hydrolases
To Be Published
|
|
6R39
 
 | |
5O3W
 
 | | Structural characterization of the fast and promiscuous macrocyclase from plant - PCY1-S562A bound to Presegetalin A1 | | Descriptor: | MAGNESIUM ION, Peptide cyclase 1, Presegetalin A1, ... | | Authors: | Ludewig, H, Czekster, C.M, Bent, A.F, Naismith, J.H. | | Deposit date: | 2017-05-25 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the Fast and Promiscuous Macrocyclase from Plant PCY1 Enables the Use of Simple Substrates.
ACS Chem. Biol., 13, 2018
|
|
6R49
 
 | | Aurora-A in complex with shape-diverse fragment 39 | | Descriptor: | (1~{S},10~{S})-12-cyclopropyl-1-oxidanyl-10-propan-2-yl-9,12-diazatricyclo[8.2.1.0^{2,7}]trideca-2(7),3,5-trien-11-one, ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, ... | | Authors: | Bayliss, R, McIntyre, P.J. | | Deposit date: | 2019-03-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Construction of a Shape-Diverse Fragment Set: Design, Synthesis and Screen against Aurora-A Kinase.
Chemistry, 25, 2019
|
|
5O7W
 
 | | Crystal structure of the 4-FLUORO RSL lectin in complex with Lewis x tetrasaccharide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Putative fucose-binding lectin protein, ... | | Authors: | Varrot, A. | | Deposit date: | 2017-06-09 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Effect of Noncanonical Amino Acids on Protein-Carbohydrate Interactions: Structure, Dynamics, and Carbohydrate Affinity of a Lectin Engineered with Fluorinated Tryptophan Analogs.
ACS Chem. Biol., 13, 2018
|
|
6REN
 
 | | Crystal structure of 3fPizza6-SH with Zn2+ | | Descriptor: | 3fPizza6-SH, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
5O32
 
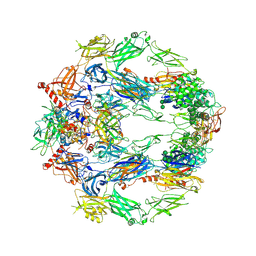 | | The structure of complement complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xue, X, Wu, J, Forneris, F, Gros, P. | | Deposit date: | 2017-05-23 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.206209 Å) | | Cite: | Regulator-dependent mechanisms of C3b processing by factor I allow differentiation of immune responses.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5O84
 
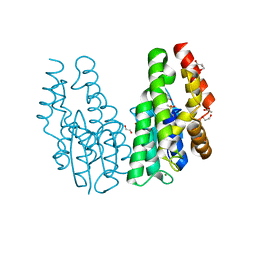 | | Glutathione S-transferase Tau 23 (partially oxidized) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FORMIC ACID, ... | | Authors: | Young, D.R, Van Molle, I, Tossounian, M, Messens, J. | | Deposit date: | 2017-06-12 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Disulfide bond formation protects Arabidopsis thaliana glutathione transferase tau 23 from oxidative damage.
Biochim. Biophys. Acta, 1862, 2018
|
|
6R3V
 
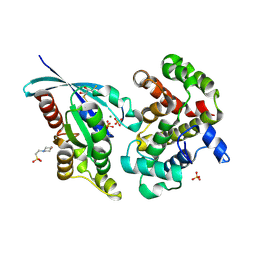 | | Crystal Structure of RhoA-GDP-Pi in Complex with RhoGAP | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jin, Y. | | Deposit date: | 2019-03-21 | | Release date: | 2019-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A GAP-GTPase-GDP-PiIntermediate Crystal Structure Analyzed by DFT Shows GTP Hydrolysis Involves Serial Proton Transfers.
Chemistry, 25, 2019
|
|
5O4T
 
 | |
6TMP
 
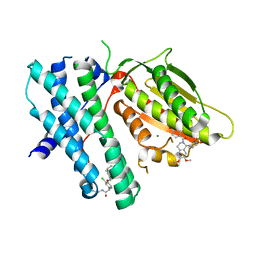 | | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures | | Descriptor: | MAGNESIUM ION, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, [2,4-bis(oxidanyl)phenyl]-[(1~{R})-6,7-dimethoxy-1-pyridin-3-yl-3,4-dihydro-1~{H}-isoquinolin-2-yl]methanone, ... | | Authors: | Baker, L.M, Aimon, A, Murray, J.B, Surgenor, A.E, Matassova, N, Roughley, S.D, von Delft, F, Hubbard, R.E. | | Deposit date: | 2019-12-05 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.076 Å) | | Cite: | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures
Commun Chem, 2020
|
|
6TAK
 
 | | Crystal structure of Escherichia coli Orotate Phosphoribosyltransferase in complex with Orotic acid and Sulfate at 1.25 Angstrom resolution | | Descriptor: | GLYCEROL, OROTIC ACID, Orotate phosphoribosyltransferase, ... | | Authors: | Navas-Yuste, S, Lopez-Estepa, M, Gomez, S, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Elucidating the Catalytic Reaction Mechanism of Orotate Phosphoribosyltransferase by Means of X-ray Crystallography and Computational Simulations
Acs Catalysis, 10, 2020
|
|
6TB0
 
 | | Crystal structure of thermostable omega transaminase 4-fold mutant from Pseudomonas jessenii | | Descriptor: | Aspartate aminotransferase family protein, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Capra, N, Rozeboom, H.J, Thunnissen, A.M.W.H, Janssen, D.B. | | Deposit date: | 2019-10-31 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Robust omega-Transaminases by Computational Stabilization of the Subunit Interface.
Acs Catalysis, 10, 2020
|
|
6TBB
 
 | | Crystal structure of S. aureus FabI in complex with NADPH and kalimantacin A (batumin) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH], Kalimantacin, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Fage, C.D, Masschelein, J. | | Deposit date: | 2019-11-01 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Kalimantacin Polyketide Antibiotics Inhibit Fatty Acid Biosynthesis in Staphylococcus aureus by Targeting the Enoyl-Acyl Carrier Protein Binding Site of FabI.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6TMU
 
 | | Crystal structure of the chaperonin gp146 from the bacteriophage EL 2 (Pseudomonas aeruginosa) in presence of ATP-BeFx, crystal form II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bracher, A, Paul, S.S, Wang, H, Wischnewski, N, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | Structure and conformational cycle of a bacteriophage-encoded chaperonin.
Plos One, 15, 2020
|
|
6TBJ
 
 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | 5-(dimethylamino)-~{N}-[6-[(2~{S},3~{R},4~{S},5~{R})-3-(hydroxymethyl)-4,5-bis(oxidanyl)piperidin-2-yl]hexyl]naphthalene-1-sulfonamide, Beta-galactosidase, putative, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2019-11-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic Insights into the Chaperoning of Human Lysosomal-Galactosidase Activity: Highly Functionalized Aminocyclopentanes and C -5a-Substituted Derivatives of 4- epi -Isofagomine.
Molecules, 25, 2020
|
|
6TC6
 
 | |
5O8Y
 
 | | Conformational dynamism for DNA interaction in Salmonella typhimurium RcsB response regulator. | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, SULFATE ION, Transcriptional regulatory protein RcsB | | Authors: | Casino, P, Marina, A, Miguel-Romero, L, Huesa, J. | | Deposit date: | 2017-06-14 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational dynamism for DNA interaction in the Salmonella RcsB response regulator.
Nucleic Acids Res., 46, 2018
|
|
5O9H
 
 | | Crystal structure of thermostabilised human C5a anaphylatoxin chemotactic receptor 1 (C5aR) in complex with NDT9513727 | | Descriptor: | 1-(1,3-benzodioxol-5-yl)-~{N}-(1,3-benzodioxol-5-ylmethyl)-~{N}-[(3-butyl-2,5-diphenyl-imidazol-4-yl)methyl]methanamine, C5a anaphylatoxin chemotactic receptor 1, CITRIC ACID, ... | | Authors: | Robertson, N, Rappas, M, Dore, A.S, Brown, J, Bottegoni, G, Koglin, M, Cansfield, J, Jazayeri, A, Cooke, R.M, Marshall, F.H. | | Deposit date: | 2017-06-19 | | Release date: | 2018-01-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the complement C5a receptor bound to the extra-helical antagonist NDT9513727.
Nature, 553, 2018
|
|
5O6E
 
 | | Structure of ScPif1 in complex with TTTGGGTT and ADP-AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase PIF1, CHLORIDE ION, ... | | Authors: | Lu, K.Y, Chen, W.F, Rety, S, Liu, N.N, Xu, X.G. | | Deposit date: | 2017-06-06 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.345 Å) | | Cite: | Insights into the structural and mechanistic basis of multifunctional S. cerevisiae Pif1p helicase.
Nucleic Acids Res., 46, 2018
|
|
5O6S
 
 | |
