2JGD
 
 | | E. COLI 2-oxoglutarate dehydrogenase (E1o) | | Descriptor: | 2-OXOGLUTARATE DEHYDROGENASE E1 COMPONENT, ADENOSINE MONOPHOSPHATE | | Authors: | Frank, R.A.W, Price, A.J, Northrop, F.D, Perham, R.N, Luisi, B.F. | | Deposit date: | 2007-02-12 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the E1 Component of the Escherichia Coli 2-Oxoglutarate Dehydrogenase Multienzyme Complex.
J.Mol.Biol., 368, 2007
|
|
4V62
 
 | | Crystal Structure of cyanobacterial Photosystem II | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Guskov, A, Gabdulkhakov, A, Kern, J, Broser, M, Zouni, A, Saenger, W. | | Deposit date: | 2008-01-17 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride
Nat.Struct.Mol.Biol., 16, 2009
|
|
4V82
 
 | | Crystal structure of cyanobacterial Photosystem II in complex with terbutryn | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Gabdulkhakov, A, Broser, M, Guskov, A, Kern, J, Glockner, C, Muh, F, Saenger, W, Zouni, A. | | Deposit date: | 2010-11-30 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of cyanobacterial photosystem II Inhibition by the herbicide terbutryn
J.Biol.Chem., 286, 2011
|
|
8CT4
 
 | | Cryo-EM structure of Mtb Lpd bound to inhibitor complex with 2-((2-cyano-N,5-dimethyl-1H-indole)-7-sulfonamido)-N-(4-(oxetan-3-yl)-3,4-dihydro-2H-benzo[b] [1,4]oxazin-7-yl)acetamide | | Descriptor: | Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, N~2~-(2-cyano-5-methyl-1H-indole-7-sulfonyl)-N~2~-methyl-N-[4-(oxetan-3-yl)-3,4-dihydro-2H-1,4-benzoxazin-7-yl]glycinamide | | Authors: | Kochanczyk, T, Arango, N, Lima, C.D. | | Deposit date: | 2022-05-13 | | Release date: | 2022-05-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Cryo-EM structure of Mtb Lpd bound to the inhibitor 2-((2-cyano-N,5-dimethyl-1H-indole)-7-sulfonamido)-N-(4-(oxetan-3-yl)-3,4-dihydro-2H-benzo[b] [1,4]oxazin-7-yl)acetamide at 2.17 Angstrom resolution
Not published
|
|
4W61
 
 | |
7KMY
 
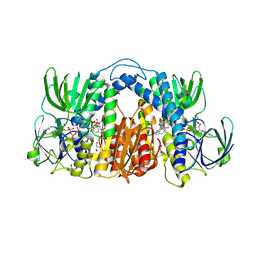 | | Structure of Mtb Lpd bound to 010705 | | Descriptor: | Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Lima, C.D. | | Deposit date: | 2020-11-03 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Whole Cell Active Inhibitors of Mycobacterial Lipoamide Dehydrogenase Afford Selectivity over the Human Enzyme through Tight Binding Interactions.
Acs Infect Dis., 7, 2021
|
|
7KPR
 
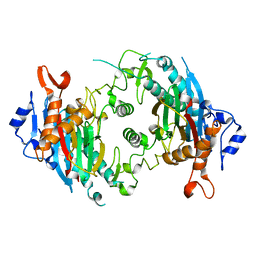 | |
8E9I
 
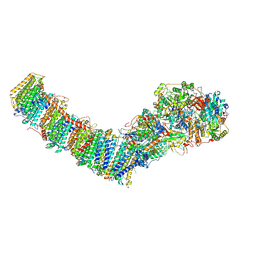 | | Mycobacterial respiratory complex I, semi-inserted quinone | | Descriptor: | (2R)-3-{[(R)-hydroxy({(1S,2R,3R,4R,5S,6S)-3,4,5-trihydroxy-2-(alpha-D-mannopyranosyloxy)-6-[(6-O-undecanoyl-beta-D-mannopyranosyl)oxy]cyclohexyl}oxy)phosphoryl]oxy}-2-(octanoyloxy)propyl undecanoate, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liang, Y, Rubinstein, J.L. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of mycobacterial respiratory complex I.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8E9H
 
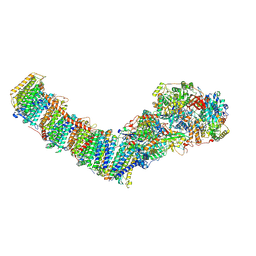 | | Mycobacterial respiratory complex I, fully-inserted quinone | | Descriptor: | (2R)-3-{[(R)-hydroxy({(1S,2R,3R,4R,5S,6S)-3,4,5-trihydroxy-2-(alpha-D-mannopyranosyloxy)-6-[(6-O-undecanoyl-beta-D-mannopyranosyl)oxy]cyclohexyl}oxy)phosphoryl]oxy}-2-(octanoyloxy)propyl undecanoate, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liang, Y, Rubinstein, J.L. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-12 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of mycobacterial respiratory complex I.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8E9G
 
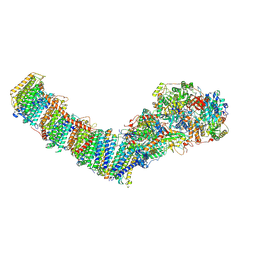 | | Mycobacterial respiratory complex I with both quinone positions modelled | | Descriptor: | (2R)-3-{[(R)-hydroxy({(1S,2R,3R,4R,5S,6S)-3,4,5-trihydroxy-2-(alpha-D-mannopyranosyloxy)-6-[(6-O-undecanoyl-beta-D-mannopyranosyl)oxy]cyclohexyl}oxy)phosphoryl]oxy}-2-(octanoyloxy)propyl undecanoate, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liang, Y, Rubinstein, J.L. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of mycobacterial respiratory complex I.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7XPI
 
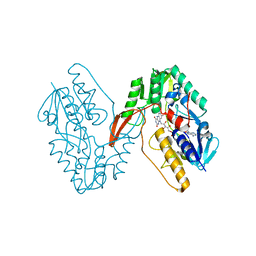 | | Structure of a oxidoreductase | | Descriptor: | Apoptosis inducing factor mitochondria associated 2, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Lv, Y, Sun, Q, Wang, Q, Zhu, D. | | Deposit date: | 2022-05-04 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into FSP1 catalysis and ferroptosis inhibition.
Nat Commun, 14, 2023
|
|
7Y11
 
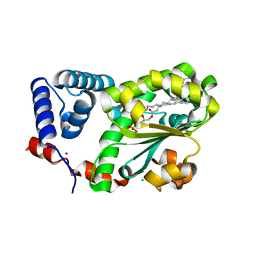 | | Crystal structure of AtSFH5-Sec14 in complex with egg PA | | Descriptor: | (2R)-1-(hexadecanoyloxy)-3-(phosphonooxy)propan-2-yl (9Z)-octadec-9-enoate, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Lu, Y.Q, Wang, X.Q, Luo, Z.P, Wu, J.W. | | Deposit date: | 2022-06-06 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Arabidopsis Sec14 proteins (SFH5 and SFH7) mediate interorganelle transport of phosphatidic acid and regulate chloroplast development.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7Y10
 
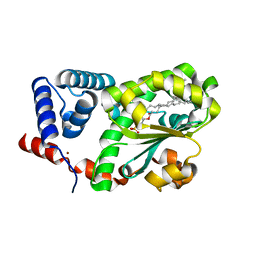 | | Crystal structure of AtSFH5-Sec14 in complex with DPPA | | Descriptor: | 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, NICKEL (II) ION, Phosphatidylinositol/phosphatidylcholine transfer protein SFH5 | | Authors: | Lu, Y.Q, Wang, X.Q, Luo, Z.P, Wu, J.W. | | Deposit date: | 2022-06-06 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Arabidopsis Sec14 proteins (SFH5 and SFH7) mediate interorganelle transport of phosphatidic acid and regulate chloroplast development.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5HEC
 
 | | CgT structure in dimer | | Descriptor: | Putative glycosyltransferase (GalT1) | | Authors: | Zhang, H, Wu, H. | | Deposit date: | 2016-01-05 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | New Helical Binding Domain Mediates a Glycosyltransferase Activity of a Bifunctional Protein.
J.Biol.Chem., 291, 2016
|
|
7YYI
 
 | |
1WCI
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, C2-1-HYDROXY-3-METHYL-BUTYL-THIAMIN, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-11-16 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
1EK4
 
 | | BETA-KETOACYL [ACYL CARRIER PROTEIN] SYNTHASE I IN COMPLEX WITH DODECANOIC ACID TO 1.85 RESOLUTION | | Descriptor: | BETA-KETOACYL [ACYL CARRIER PROTEIN] SYNTHASE I, LAURIC ACID | | Authors: | Olsen, J.G, Kadziola, A, Siggaard-Andersen, M, von Wettstein-Knowles, P, Larsen, S. | | Deposit date: | 2000-03-06 | | Release date: | 2001-04-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of beta-ketoacyl-acyl carrier protein synthase I complexed with fatty acids elucidate its catalytic machinery.
Structure, 9, 2001
|
|
1EPS
 
 | | STRUCTURE AND TOPOLOGICAL SYMMETRY OF THE GLYPHOSPHATE 5-ENOL-PYRUVYLSHIKIMATE-3-PHOSPHATE SYNTHASE: A DISTINCTIVE PROTEIN FOLD | | Descriptor: | 5-ENOL-PYRUVYL-3-PHOSPHATE SYNTHASE | | Authors: | Stallings, W.C, Abdel-Meguid, S.S, Lim, L.W, Shieh, H.-S, Dayringer, H.E, Leimgruber, N.K, Stegeman, R.A, Anderson, K.S, Sikorski, J.A, Padgette, S.R, Kishore, G.M. | | Deposit date: | 1991-04-05 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and topological symmetry of the glyphosate target 5-enolpyruvylshikimate-3-phosphate synthase: a distinctive protein fold.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1EV8
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CYS167 MUTANT OF ESCHERICHIA COLI | | Descriptor: | THYMIDYLATE SYNTHASE | | Authors: | Phan, J, Mahdavian, E, Nivens, M.C, Minor, W, Berger, S, Spencer, H.T, Dunlap, R.B, Lebioda, L. | | Deposit date: | 2000-04-19 | | Release date: | 2000-05-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalytic cysteine of thymidylate synthase is activated upon substrate binding.
Biochemistry, 39, 2000
|
|
1EVF
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CYS167 MUTANT OF ESCHERICHIA COLI | | Descriptor: | SULFATE ION, THYMIDYLATE SYNTHASE | | Authors: | Phan, J, Mahdavian, E, Nivens, M.C, Minor, W, Berger, S, Spencer, H.T, Dunlap, R.B, Lebioda, L. | | Deposit date: | 2000-04-19 | | Release date: | 2000-05-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic cysteine of thymidylate synthase is activated upon substrate binding.
Biochemistry, 39, 2000
|
|
1EJC
 
 | | Crystal structure of unliganded mura (type2) | | Descriptor: | GLYCEROL, PHOSPHATE ION, UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYLTRANSFERASE | | Authors: | Eschenburg, S, Schonbrunn, E. | | Deposit date: | 2000-03-02 | | Release date: | 2000-10-25 | | Last modified: | 2011-07-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparative X-ray analysis of the un-liganded fosfomycin-target murA.
Proteins, 40, 2000
|
|
1EYS
 
 | | CRYSTAL STRUCTURE OF PHOTOSYNTHETIC REACTION CENTER FROM A THERMOPHILIC BACTERIUM, THERMOCHROMATIUM TEPIDUM | | Descriptor: | 2-O-octyl-beta-D-glucopyranose, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Nogi, T, Fathir, I, Kobayashi, M, Nozawa, T, Miki, K. | | Deposit date: | 2000-05-08 | | Release date: | 2000-12-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of photosynthetic reaction center and high-potential iron-sulfur protein from Thermochromatium tepidum: thermostability and electron transfer.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1OLS
 
 | | Roles of His291-alpha and His146-beta' in the reductive acylation reaction catalyzed by human branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, GLYCEROL, ... | | Authors: | Wynn, R.M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2003-08-12 | | Release date: | 2003-08-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Roles of His291-Alpha and His146-Beta' in the Reductive Acylation Reaction Catalyzed by Human Branched-Chain Alpha-Ketoacid Dehydrogenase: Refined Phosphorylation Loop Structure in the Active Site.
J.Biol.Chem., 278, 2003
|
|
3II4
 
 | |
6VEF
 
 | |
