2HGC
 
 | | Solution NMR structure of the YjcQ protein from Bacillus subtilis. Northeast Structural Genomics target SR346. | | Descriptor: | YjcQ protein | | Authors: | Rossi, P, Cort, J.R, Ho, C.K, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-26 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the YjcQ protein from Bacillus subtilis. Northeast Structural Genomics target SR346. (CASP Target)
To be Published
|
|
2LEB
 
 | | Solution structure of human SRSF2 (SC35) RRM in complex with 5'-UCCAGU-3' | | Descriptor: | RNA (5'-R(*UP*CP*CP*AP*GP*U)-3'), Serine/arginine-rich splicing factor 2 | | Authors: | Daubner, G.M, Clery, A, Jayne, S, Stevenin, J, Allain, F.H.-T. | | Deposit date: | 2011-06-15 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well.
Embo J., 31, 2012
|
|
2LEA
 
 | | Solution structure of human SRSF2 (SC35) RRM | | Descriptor: | Serine/arginine-rich splicing factor 2 | | Authors: | Daubner, G.M, Clery, A, Jayne, S, Stevenin, J, Allain, F.H.-T. | | Deposit date: | 2011-06-15 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well.
Embo J., 31, 2012
|
|
2JOM
 
 | |
2M8O
 
 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in DPC | | Descriptor: | Transmembrane protein gp41 | | Authors: | Serrano, S, Huarte, N, Nieva, J.L, Jimenez, M. | | Deposit date: | 2013-05-23 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Immunogenicity of a Peptide Vaccine, Including the Complete HIV-1 gp41 2F5 Epitope: IMPLICATIONS FOR ANTIBODY RECOGNITION MECHANISM AND IMMUNOGEN DESIGN.
J.Biol.Chem., 289, 2014
|
|
6LZ9
 
 | |
1IIC
 
 | |
6MUJ
 
 | | Formylglycine generating enzyme bound to copper | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Lafrance-Vanasse, J, Appel, M.J, Tsai, C.-L, Bertozzi, C, Tainer, J.A. | | Deposit date: | 2018-10-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Formylglycine-generating enzyme binds substrate directly at a mononuclear Cu(I) center to initiate O2activation.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3J9Z
 
 | | Activation of GTP Hydrolysis in mRNA-tRNA Translocation by Elongation Factor G | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Li, W, Liu, Z, Koripella, R.K, Langlois, R, Sanyal, S, Frank, J. | | Deposit date: | 2015-03-27 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of GTP hydrolysis in mRNA-tRNA translocation by elongation factor G.
Sci Adv, 1, 2015
|
|
3JA1
 
 | | Activation of GTP Hydrolysis in mRNA-tRNA Translocation by Elongation Factor G | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Li, W, Liu, Z, Koripella, R.K, Langlois, R, Sanyal, S, Frank, J. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of GTP hydrolysis in mRNA-tRNA translocation by elongation factor G.
Sci Adv, 1, 2015
|
|
1JFH
 
 | |
1LGL
 
 | | Solution structure of HERG-specific scorpion toxin BeKm-1 | | Descriptor: | BeKm-1 toxin | | Authors: | Korolokova, Y.V, Bocharov, E.V, Angelo, K, Maslennikov, I.V, Grinenko, O.V, Lipkin, A.V, Nosireva, E.D, Pluzhnikov, K.A, Olesen, S.-P, Arseniev, A.S, Grishin, E.V. | | Deposit date: | 2002-04-16 | | Release date: | 2002-11-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | New binding site on common molecular scaffold provides HERG channel specificity of scorpion toxin BeKm-1.
J.Biol.Chem., 277, 2002
|
|
1LO3
 
 | | Retro-Diels-Alderase Catalytic Antibody: Product Analogue | | Descriptor: | 3-(10-METHYL-ANTHRACEN-9-YL)-PROPIONIC ACID, If kappa light chain, Ig gamma 2a heavy chain | | Authors: | Hugot, M, Reymond, J.L, Baumann, U. | | Deposit date: | 2002-05-06 | | Release date: | 2002-07-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural basis for the activity of retro-Diels-Alder catalytic antibodies: evidence for a catalytic aromatic residue.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1IID
 
 | | Crystal Structure of Saccharomyces cerevisiae N-myristoyltransferase with Bound S-(2-oxo)pentadecylCoA and the Octapeptide GLYASKLA | | Descriptor: | NICKEL (II) ION, Octapeptide GLYASKLA, Peptide N-myristoyltransferase, ... | | Authors: | Farazi, T.A, Gordon, J.I, Waksman, G. | | Deposit date: | 2001-04-22 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Saccharomyces cerevisiae N-myristoyltransferase with bound myristoylCoA and peptide provide insights about substrate recognition and catalysis.
Biochemistry, 40, 2001
|
|
1J5J
 
 | | Solution structure of HERG-specific scorpion toxin BeKm-1 | | Descriptor: | BeKm-1 toxin | | Authors: | Korolokova, Y.V, Bocharov, E.V, Angelo, K, Maslennikov, I.V, Grinenko, O.V, Lipkin, A.V, Nosireva, E.D, Pluzhnikov, K.A, Olesen, S.-P, Arseniev, A.S, Grishin, E.V. | | Deposit date: | 2002-04-16 | | Release date: | 2002-11-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | New binding site on common molecular scaffold provides HERG channel specificity of scorpion toxin BeKm-1.
J.Biol.Chem., 277, 2002
|
|
1LO2
 
 | | Retro-Diels-Alderase Catalytic Antibody | | Descriptor: | If kappa light chain, Ig gamma 2a heavy chain, [2'-CARBOXYLETHYL]-10-METHYL-ANTHRACENE ENDOPEROXIDE | | Authors: | Hugot, M, Reymond, J.L, Baumann, U. | | Deposit date: | 2002-05-06 | | Release date: | 2002-06-26 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural basis for the activity of retro-Diels-Alder catalytic antibodies: evidence for a catalytic aromatic residue.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LO4
 
 | | Retro-Diels-Alderase Catalytic antibody 9D9 | | Descriptor: | If kappa light chain, Ig gamma 2a heavy chain | | Authors: | Hugot, M, Reymond, J.L, Baumann, U. | | Deposit date: | 2002-05-06 | | Release date: | 2002-07-03 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A structural basis for the activity of retro-Diels-Alder catalytic antibodies: evidence for a catalytic aromatic residue.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LNS
 
 | |
3LN0
 
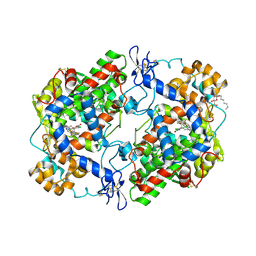 | | Structure of compound 5c-S bound at the active site of COX-2 | | Descriptor: | (2S)-6,8-dichloro-2-(trifluoromethyl)-2H-chromene-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kiefer, J.R, Kurumbail, R.G, Stallings, W.C, Pawlitz, J.L. | | Deposit date: | 2010-02-01 | | Release date: | 2010-10-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The novel benzopyran class of selective cyclooxygenase-2 inhibitors. Part 2: The second clinical candidate having a shorter and favorable human half-life.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1LO0
 
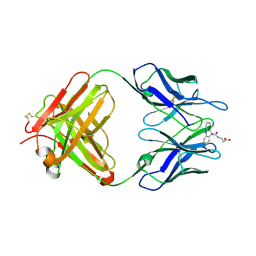 | | Catalytic Retro-Diels-Alderase Transition State Analogue Complex | | Descriptor: | 3-{[(9-CYANO-9,10-DIHYDRO-10-METHYLACRIDIN-9-YL)CARBONYL]AMINO}PROPANOIC ACID, If kappa light chain, Ig gamma 2a heavy chain | | Authors: | Hugot, M, Reymond, J.L, Baumann, U. | | Deposit date: | 2002-05-05 | | Release date: | 2002-06-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural basis for the activity of retro-Diels-Alder catalytic antibodies: evidence for a catalytic aromatic residue.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1J5S
 
 | |
1M39
 
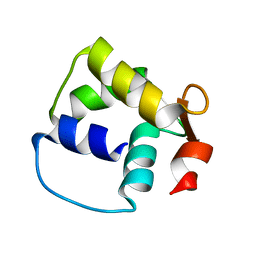 | | Solution structure of the C-terminal fragment (F86-I165) of the human centrin 2 in calcium saturated form | | Descriptor: | Caltractin, isoform 1 | | Authors: | Matei, E, Miron, S, Blouquit, Y, Duchambon, P, Durussel, P, Cox, J.A, Craescu, C.T. | | Deposit date: | 2002-06-27 | | Release date: | 2003-03-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | C-terminal half of human centrin 2 behaves like a regulatory EF-hand domain
Biochemistry, 42, 2003
|
|
8XXN
 
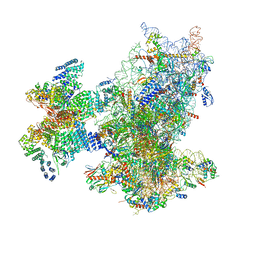 | | Cryo-EM structure of the human 43S ribosome with PDCD4 | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 34, 2024
|
|
8V61
 
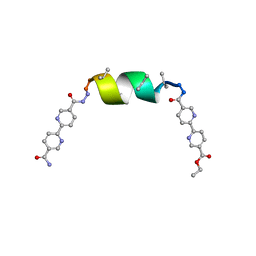 | | UIC-1 mutant - UIC-1-L6I | | Descriptor: | UIC-1-L6I | | Authors: | Heinz-Kunert, S.L. | | Deposit date: | 2023-12-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Pore Restructuring of Peptide Frameworks by Mutations at Distal Packing Residues.
Biomacromolecules, 25, 2024
|
|
8V5X
 
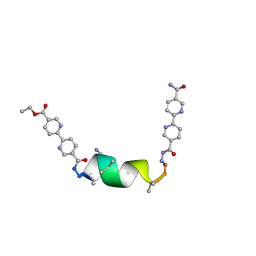 | | UIC-1 mutant - UIC-1-L6A | | Descriptor: | ACETONITRILE, UIC-1-L6A | | Authors: | Heinz-Kunert, S.L. | | Deposit date: | 2023-12-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Pore Restructuring of Peptide Frameworks by Mutations at Distal Packing Residues.
Biomacromolecules, 25, 2024
|
|
