7JFY
 
 | | GAS41 YEATS domain in complex with 5 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, N-(5-{3-[(2S)-1,3-thiazolidin-2-yl]azetidine-1-carbonyl}thiophen-2-yl)-L-prolinamide, ... | | Authors: | Linhares, B.M, Listunov, D, Winkler, A, Grembecka, J, Cierpicki, T. | | Deposit date: | 2020-07-17 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.100557 Å) | | Cite: | GAS41 YEATS domain in complex with 5
To Be Published
|
|
7JKY
 
 | | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with YF3-126 | | Descriptor: | Bromodomain-containing protein 4, N-(1-[1,1-di(pyridin-2-yl)ethyl]-6-{1-methyl-6-oxo-5-[(piperidin-4-yl)amino]-1,6-dihydropyridin-3-yl}-1H-indol-4-yl)ethanesulfonamide, SODIUM ION | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | Deposit date: | 2020-07-29 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Novel Pyrrolopyridone Bromodomain and Extra-Terminal Motif (BET) Inhibitors Effective in Endocrine-Resistant ER+ Breast Cancer with Acquired Resistance to Fulvestrant and Palbociclib.
J.Med.Chem., 63, 2020
|
|
7JKW
 
 | | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with ZN1-99 | | Descriptor: | Bromodomain-containing protein 4, N-(6-{5-[(azetidin-3-yl)amino]-1-methyl-6-oxo-1,6-dihydropyridin-3-yl}-1-[1,1-di(pyridin-2-yl)ethyl]-1H-indol-4-yl)ethanesulfonamide | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | Deposit date: | 2020-07-29 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Novel Pyrrolopyridone Bromodomain and Extra-Terminal Motif (BET) Inhibitors Effective in Endocrine-Resistant ER+ Breast Cancer with Acquired Resistance to Fulvestrant and Palbociclib.
J.Med.Chem., 63, 2020
|
|
7JKX
 
 | | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with YF3-6 | | Descriptor: | Bromodomain-containing protein 4, N-{1-[1,1-di(pyridin-2-yl)ethyl]-6-[1-methyl-5-(methylamino)-6-oxo-1,6-dihydropyridin-3-yl]-1H-indol-4-yl}ethanesulfonamide | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | Deposit date: | 2020-07-29 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with YF3-6
To Be Published
|
|
2XD7
 
 | | Crystal structure of the macro domain of human core histone H2A | | Descriptor: | CORE HISTONE MACRO-H2A.2 | | Authors: | Vollmar, M, Phillips, C, Carpenter, E.P, Muniz, J.R.C, Krojer, T, Ugochukwu, E, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Gileadi, O. | | Deposit date: | 2010-04-29 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of the Macro Domain of Human Core Histone H2A
To be Published
|
|
2XNE
 
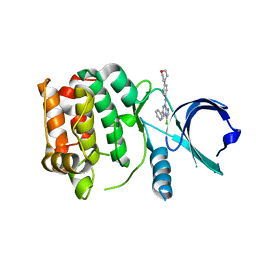 | | Structure of Aurora-A bound to an imidazopyrazine inhibitor | | Descriptor: | 3-chloro-N-(4-morpholin-4-ylphenyl)-6-pyridin-3-ylimidazo[1,2-a]pyrazin-8-amine, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2010-08-02 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of imidazo[1,2-a]pyrazine derivatives as selective inhibitors of Aurora-A kinase in cells.
Bioorg. Med. Chem. Lett., 20, 2010
|
|
4O3M
 
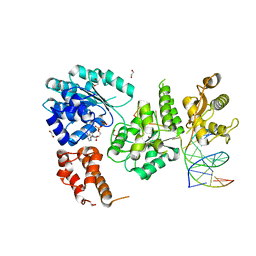 | | Ternary complex of Bloom's syndrome helicase | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*AP*GP*CP*GP*TP*CP*GP*AP*GP*AP*TP*CP*CP*AP*AP*G)-3', 5'-D(*CP*TP*TP*GP*GP*AP*TP*CP*TP*CP*GP*AP*CP*GP*CP*TP*CP*TP*CP*CP*CP*TP*TP*A)-3', ... | | Authors: | Swan, M.K, Bertrand, J. | | Deposit date: | 2013-12-18 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of human Bloom's syndrome helicase in complex with ADP and duplex DNA.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4O63
 
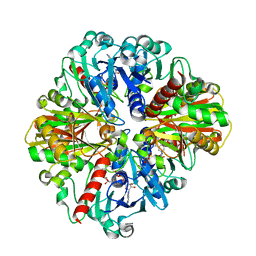 | | Co-enzyme Induced Conformational Changes in Bovine Eye Glyceraldehyde 3-Phosphate Dehydrogenase | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Baker, B.Y, Shi, W, Wang, B, Palczewski, K. | | Deposit date: | 2013-12-20 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | High-resolution crystal structures of the photoreceptor glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with three and four-bound NAD molecules.
Protein Sci., 23, 2014
|
|
4O70
 
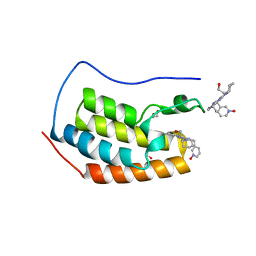 | | Crystal structure of the first bromodomain of human BRD4 in complex with DINACICLIB | | Descriptor: | 1,2-ETHANEDIOL, 3-[({3-ethyl-5-[(2S)-2-(2-hydroxyethyl)piperidin-1-yl]pyrazolo[1,5-a]pyrimidin-7-yl}amino)methyl]-1-hydroxypyridinium, Bromodomain-containing protein 4 | | Authors: | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
4O7F
 
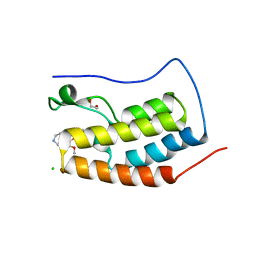 | | Crystal structure of the first bromodomain of human BRD4 in complex with SB-251527 | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-(4-fluorophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]-2-(2-methoxyphenoxy)pyrimidine, Bromodomain-containing protein 4, ... | | Authors: | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
4O72
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with NU7441 | | Descriptor: | 1,2-ETHANEDIOL, 8-(dibenzo[b,d]thiophen-4-yl)-2-(morpholin-4-yl)-4H-chromen-4-one, Bromodomain-containing protein 4, ... | | Authors: | Zhu, J.-Y, Ember, S.W, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
2UXX
 
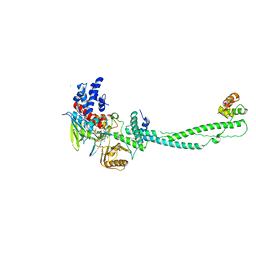 | | Human LSD1 Histone Demethylase-CoREST in complex with an FAD- tranylcypromine adduct | | Descriptor: | CHLORIDE ION, FAD-trans-2-Phenylcyclopropylamine Adduct, GLYCEROL, ... | | Authors: | Yang, M, Culhane, J.C, Machius, M, Cole, P.A, Yu, H. | | Deposit date: | 2007-03-30 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural Basis for the Inhibition of the Lsd1 Histone Demethylase by the Antidepressant Trans-2-Phenylcyclopropylamine.
Biochemistry, 46, 2007
|
|
2VY4
 
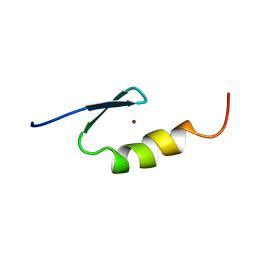 | | U11-48K CHHC ZN-FINGER DOMAIN | | Descriptor: | U11/U12 SMALL NUCLEAR RIBONUCLEOPROTEIN 48 KDA PROTEIN, ZINC ION | | Authors: | Tidow, H, Andreeva, A, Rutherford, T.J, Fersht, A.R. | | Deposit date: | 2008-07-17 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the U11-48K CHHC zinc-finger domain that specifically binds the 5' splice site of U12-type introns.
Structure, 17, 2009
|
|
4O7A
 
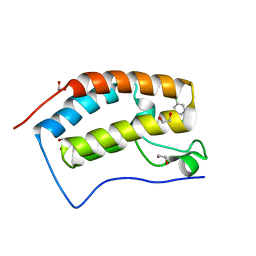 | | Crystal structure of the first bromodomain of human BRD4 in complex with SB-409514 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-chloro-4-hydroxyphenyl)amino]-4-(3-chlorophenyl)-1H-pyrrole-2,5-dione, Bromodomain-containing protein 4 | | Authors: | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
4O7C
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SB-614067-R | | Descriptor: | 1,2-ETHANEDIOL, 4-[(5Z)-5-(1-nitroso-2,3-dihydro-5H-inden-5-ylidene)-2-(piperidin-4-yl)-3,5-dihydro-4H-imidazol-4-ylidene]-1,4-dihydropyridine, Bromodomain-containing protein 4 | | Authors: | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
2XNG
 
 | | Structure of Aurora-A bound to a selective imidazopyrazine inhibitor | | Descriptor: | N-(3-{3-chloro-8-[(4-morpholin-4-ylphenyl)amino]imidazo[1,2-a]pyrazin-6-yl}benzyl)methanesulfonamide, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2010-08-02 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structure-based design of imidazo[1,2-a]pyrazine derivatives as selective inhibitors of Aurora-A kinase in cells.
Bioorg. Med. Chem. Lett., 20, 2010
|
|
2XO3
 
 | |
2XGY
 
 | | Complex of Rabbit Endogenous Lentivirus (RELIK)Capsid with Cyclophilin A | | Descriptor: | GLYCEROL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A, RELIK CAPSID N-TERMINAL DOMAIN | | Authors: | Goldstone, D.C, Robertson, L.E, Haire, L.F, Stoye, J.P, Taylor, I.A. | | Deposit date: | 2010-06-08 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Analysis of Prehistoric Lentiviruses Uncovers an Ancient Molecular Interface.
Cell Host Microbe, 8, 2010
|
|
2YAC
 
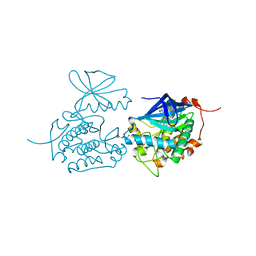 | | Crystal structure of Polo-like kinase 1 in complex with NMS-P937 | | Descriptor: | 1-(2-HYDROXYETHYL)-8-[[5-(4-METHYLPIPERAZIN-1-YL)-2-(TRIFLUOROMETHOXY)PHENYL]AMINO]-4,5-DIHYDROPYRIMIDO[5,4-G]INDAZOLE-3-CARBOXAMIDE, L(+)-TARTARIC ACID, SERINE/THREONINE-PROTEIN KINASE PLK1, ... | | Authors: | Bertrand, J.A, Bossi, R.T. | | Deposit date: | 2011-02-18 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nms-P937, a 4,5-Dihydro-1H-Pyrazolo[4,3-H]Quinazoline Derivative as Potent and Selective Polo-Like Kinase 1 Inhibitor.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3DFP
 
 | |
7MNX
 
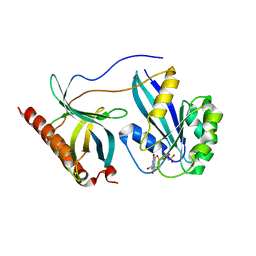 | | Crystal Structure of Nup358/RanBP2 Ran-binding domain 2 in complex with Ran-GPPNHP | | Descriptor: | E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, MAGNESIUM ION, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MO5
 
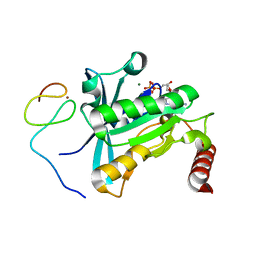 | | Crystal Structure of the ZnF4 of Nucleoporin NUP153 in complex with Ran-GDP | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNS
 
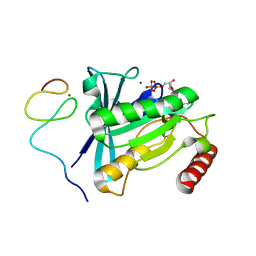 | | Crystal Structure of the ZnF4 of Nucleoporin NUP358/RanBP2 in complex with Ran-GDP | | Descriptor: | E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNV
 
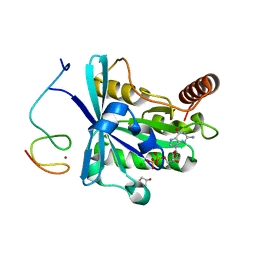 | | Crystal Structure of the ZnF8 of Nucleoporin NUP358/RanBP2 in complex with Ran-GDP | | Descriptor: | E3 SUMO-protein ligase RanBP2, GLYCEROL, GTP-binding nuclear protein Ran, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MO4
 
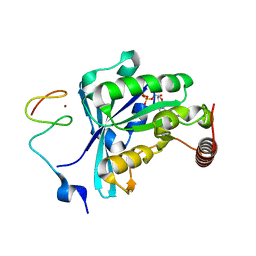 | | Crystal Structure of the ZnF3 of Nucleoporin NUP153 in complex with Ran-GDP, resolution 2.4 Angstrom | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
