6BMI
 
 | | Crystal Structure of GltPh R397C in complex with L-Serine | | Descriptor: | Glutamate transporter homolog, SERINE, SODIUM ION | | Authors: | Font, J, Scopelliti, A.J, Vandenberg, R.J, Boudker, O, Ryan, R.M. | | Deposit date: | 2017-11-14 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural characterisation reveals insights into substrate recognition by the glutamine transporter ASCT2/SLC1A5.
Nat Commun, 9, 2018
|
|
1EEH
 
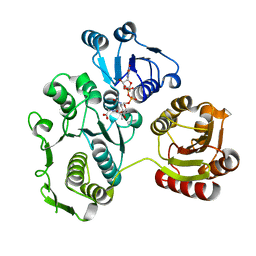 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE, URIDINE-5'-DIPHOSPHATE-N-ACETYLMURAMOYL-L-ALANINE | | Authors: | Bertrand, J.A, Fanchon, E, Martin, L, Chantalat, L, Auger, G, Blanot, D, van Heijenoort, J, Dideberg, O. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | "Open" structures of MurD: domain movements and structural similarities with folylpolyglutamate synthetase.
J.Mol.Biol., 301, 2000
|
|
2H8D
 
 | | Crystal structure of deoxy hemoglobin from Trematomus bernacchii at pH 8.4 | | Descriptor: | Hemoglobin alpha subunit, Hemoglobin beta subunit, POTASSIUM ION, ... | | Authors: | Mazzarella, L, Vergara, A, Vitagliano, L, Merlino, A, Bonomi, G, Scala, S, Verde, C, di Prisco, G. | | Deposit date: | 2006-06-07 | | Release date: | 2006-08-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | High resolution crystal structure of deoxy hemoglobin from Trematomus bernacchii at different pH values: The role of histidine residues in modulating the strength of the root effect.
Proteins, 65, 2006
|
|
2PTL
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF AN IMMUNOGLOBULIN LIGHT CHAIN-BINDING DOMAIN OF PROTEIN L. COMPARISON WITH THE IGG-BINDING DOMAINS OF PROTEIN G | | Descriptor: | PROTEIN L | | Authors: | Wikstroem, M, Drakenberg, T, Forsen, S, Sjoebring, U, Bjoerck, L. | | Deposit date: | 1994-08-12 | | Release date: | 1994-10-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of an immunoglobulin light chain-binding domain of protein L. Comparison with the IgG-binding domains of protein G.
Biochemistry, 33, 1994
|
|
1HW4
 
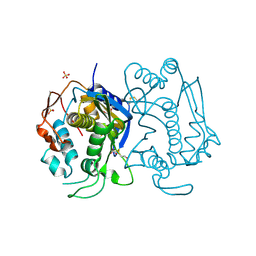 | | STRUCTURE OF THYMIDYLATE SYNTHASE SUGGESTS ADVANTAGES OF CHEMOTHERAPY WITH NONCOMPETITIVE INHIBITORS | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, THYMIDYLATE SYNTHASE | | Authors: | Phan, J, Steadman, J.D, Koli, S, Ding, W.C, Minor, W, Dunlap, R.B, Berger, S.H, Lebioda, L. | | Deposit date: | 2001-01-09 | | Release date: | 2001-01-24 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure of human thymidylate synthase suggests advantages of chemotherapy with noncompetitive inhibitors.
J.Biol.Chem., 276, 2001
|
|
2F1V
 
 | | Outer membrane protein OmpW | | Descriptor: | GLYCEROL, Outer membrane protein W | | Authors: | van den Berg, B. | | Deposit date: | 2005-11-15 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The outer membrane protein OmpW forms an eight-stranded beta-barrel with a hydrophobic channel.
J.Biol.Chem., 281, 2006
|
|
2GU2
 
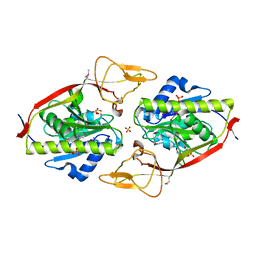 | | Crystal Structure of an Aspartoacylase from Rattus norvegicus | | Descriptor: | Aspa protein, SULFATE ION, ZINC ION | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-04-28 | | Release date: | 2006-06-20 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structure of aspartoacylase, the brain enzyme impaired in Canavan disease.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
8W7S
 
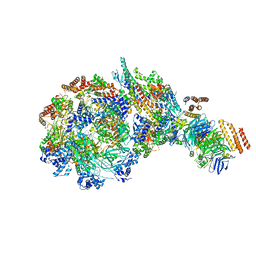 | | Yeast replisome in state IV | | Descriptor: | Cell division control protein 45, DNA (71-mer), DNA polymerase alpha-binding protein, ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D. | | Deposit date: | 2023-08-31 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (7.39 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
8KG8
 
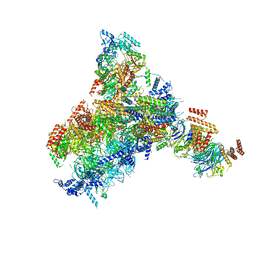 | | Yeast replisome in state II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, DNA (61-mer), ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | Deposit date: | 2023-08-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
8KG9
 
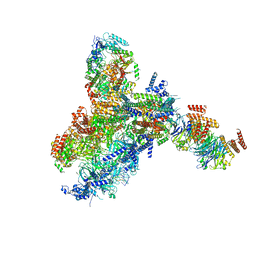 | | Yeast replisome in state III | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, DNA (61-mer), ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | Deposit date: | 2023-08-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.52 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
8KG6
 
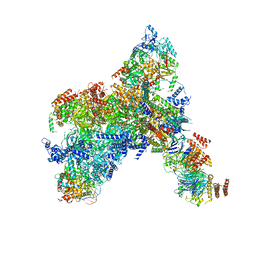 | | Yeast replisome in state I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, Chromosome segregation in meiosis protein 3, ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | Deposit date: | 2023-08-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
7CBT
 
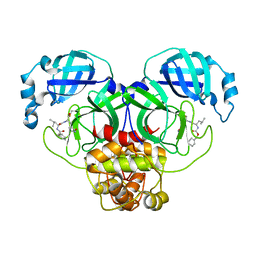 | | The crystal structure of SARS-CoV-2 main protease in complex with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Shi, Y, Peng, G. | | Deposit date: | 2020-06-13 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.346 Å) | | Cite: | The preclinical inhibitor GS441524 in combination with GC376 efficaciously inhibited the proliferation of SARS-CoV-2 in the mouse respiratory tract.
Emerg Microbes Infect, 10, 2021
|
|
1J85
 
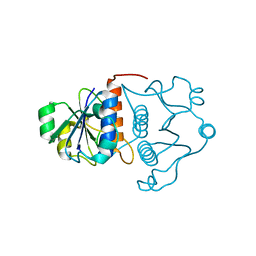 | | Structure of YibK from Haemophilus influenzae (HI0766), a truncated sequence homolog of tRNA (guanosine-2'-O-) methyltransferase (SpoU) | | Descriptor: | YibK | | Authors: | Lim, K, Zhang, H, Toedt, J, Tempcyzk, A, Krajewski, W, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-20 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the YibK methyltransferase from Haemophilus influenzae
(HI0766): A cofactor bound at a site formed by a knot
Proteins, 51, 2003
|
|
1MH3
 
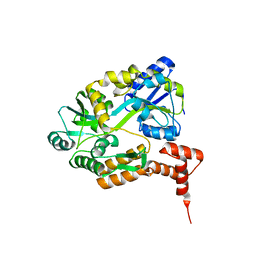 | | maltose binding-a1 homeodomain protein chimera, crystal form I | | Descriptor: | maltose binding-a1 homeodomain protein chimera | | Authors: | Ke, A, Wolberger, C. | | Deposit date: | 2002-08-19 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into binding cooperativity of MATa1/MATalpha2 from the crystal structure of a MATa1 homeodomain-maltose binding protein chimera
Protein Sci., 12, 2003
|
|
1NIK
 
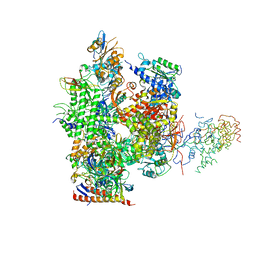 | | Wild Type RNA Polymerase II | | Descriptor: | DNA-directed RNA polymerase I, II and III 23 kDa polypeptide, DNA-directed RNA polymerase II, ... | | Authors: | Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2002-12-24 | | Release date: | 2003-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Complete, 12-subunit RNA Polymerase II at 4.1-A resolution: implications for the initiation of transcription.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1MH4
 
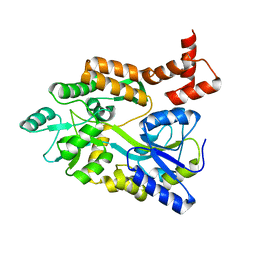 | | maltose binding-a1 homeodomain protein chimera, crystal form II | | Descriptor: | maltose binding-a1 homeodomain protein chimera | | Authors: | Ke, A, Wolberger, C. | | Deposit date: | 2002-08-19 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into binding cooperativity of MATa1/MATalpha2 from the crystal structure of a MATa1 homeodomain-maltose binding protein chimera
Protein Sci., 12, 2003
|
|
1DYW
 
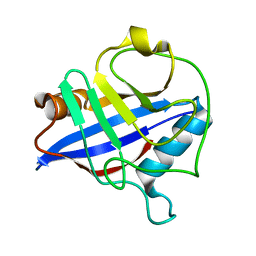 | | Biochemical and structural characterization of a divergent loop cyclophilin from Caenorhabditis elegans | | Descriptor: | CYCLOPHILIN 3 | | Authors: | Dornan, J, Page, A.P, Taylor, P, Wu, S.Y, Winter, A.D, Husi, H, Walkinshaw, M.D. | | Deposit date: | 2000-02-10 | | Release date: | 2000-06-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical and Structural Characterization of a Divergent Loop Cyclophilin from Caenorhabditis Elegans
J.Biol.Chem., 274, 1999
|
|
7D7S
 
 | |
2ISW
 
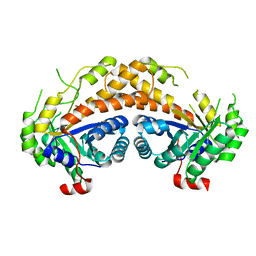 | | Structure of Giardia fructose-1,6-biphosphate aldolase in complex with phosphoglycolohydroxamate | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, Putative fructose-1,6-bisphosphate aldolase, ZINC ION | | Authors: | Galkin, A, Herzberg, O. | | Deposit date: | 2006-10-18 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization, kinetics, and crystal structures of fructose-1,6-bisphosphate aldolase from the human parasite, Giardia lamblia.
J.Biol.Chem., 282, 2007
|
|
8PFK
 
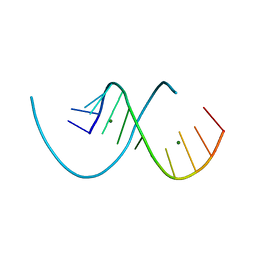 | | RNA structure with 1-methylpseudoridine, C2 space group | | Descriptor: | MAGNESIUM ION, RNA (12-mer) | | Authors: | Spingler, B, McAuley, K, Nievergelt, P, Thorn, A. | | Deposit date: | 2023-06-16 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.321 Å) | | Cite: | RNA oligomers at atomic resolution containing 1-methylpseudouridine, an essential building block of mRNA vaccines.
Chemmedchem, 19, 2024
|
|
8PFQ
 
 | | RNA structure with 1-methylpseudoridine, P1 space group | | Descriptor: | Chains: A,B, MAGNESIUM ION | | Authors: | Spingler, B, McAuley, K, Nievergelt, P, Thorn, A. | | Deposit date: | 2023-06-16 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | RNA oligomers at atomic resolution containing 1-methylpseudouridine, an essential building block of mRNA vaccines.
Chemmedchem, 19, 2024
|
|
1ORT
 
 | | ORNITHINE TRANSCARBAMOYLASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | ORNITHINE TRANSCARBAMOYLASE | | Authors: | Villeret, V, Dideberg, O. | | Deposit date: | 1995-08-24 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Pseudomonas aeruginosa catabolic ornithine transcarbamoylase at 3.0-A resolution: a different oligomeric organization in the transcarbamoylase family.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1FSW
 
 | | AMPC BETA-LACTAMASE FROM E. COLI COMPLEXED WITH INHIBITOR CEPHALOTHINBORONIC ACID | | Descriptor: | CEPHALOSPORINASE, N-2-THIOPHEN-2-YL-ACETAMIDE BORONIC ACID, PHOSPHATE ION | | Authors: | Caselli, E, Powers, R.A, Blaszczak, L.C, Wu, C.Y, Prati, F, Shoichet, B.K. | | Deposit date: | 2000-09-11 | | Release date: | 2001-03-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Energetic, structural, and antimicrobial analyses of beta-lactam side chain recognition by beta-lactamases.
Chem.Biol., 8, 2001
|
|
2VDB
 
 | | Structure of human serum albumin with S-naproxen and the GA module | | Descriptor: | (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid, DECANOIC ACID, PEPTOSTREPTOCOCCAL ALBUMIN-BINDING PROTEIN, ... | | Authors: | Lejon, S, Cramer, J.F, Nordberg, P.A. | | Deposit date: | 2007-10-04 | | Release date: | 2008-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural Basis for the Binding of Naproxen to Human Serum Albumin in the Presence of Fatty Acids and the Ga Module.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2ISV
 
 | | Structure of Giardia fructose-1,6-biphosphate aldolase in complex with phosphoglycolohydroxamate | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, Putative fructose-1,6-bisphosphate aldolase, ZINC ION | | Authors: | Galkin, A, Herzberg, O. | | Deposit date: | 2006-10-18 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization, kinetics, and crystal structures of fructose-1,6-bisphosphate aldolase from the human parasite, Giardia lamblia.
J.Biol.Chem., 282, 2007
|
|
