6RTE
 
 | | Dihydro-heme d1 dehydrogenase NirN in complex with DHE | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cytochrome c, HEME C | | Authors: | Kluenemann, T, Preuss, A, Layer, G, Blankenfeldt, W. | | Deposit date: | 2019-05-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Dihydro-Heme d1Dehydrogenase NirN from Pseudomonas aeruginosa Reveals Amino Acid Residues Essential for Catalysis.
J.Mol.Biol., 431, 2019
|
|
5MYV
 
 | | Crystal structure of SRPK2 in complex with compound 1 | | Descriptor: | 5-methyl-~{N}-[2-(4-methylpiperazin-1-yl)-5-(trifluoromethyl)phenyl]furan-2-carboxamide, DIMETHYL SULFOXIDE, SRSF protein kinase 2,SRSF protein kinase 2, ... | | Authors: | Chaikuad, A, Pike, A.C.W, Savitsky, P, von Delft, F, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-29 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Development of Potent, Selective SRPK1 Inhibitors as Potential Topical Therapeutics for Neovascular Eye Disease.
ACS Chem. Biol., 12, 2017
|
|
5N6U
 
 | |
5MZJ
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with theophylline at 2.0A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Cheng, R.K.Y, Segala, E, Robertson, N, Deflorian, F, Dore, A.S, Errey, J.C, Fiez-Vandal, C, Marshall, F.H, Cooke, R.M. | | Deposit date: | 2017-01-31 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity.
Structure, 25, 2017
|
|
5MZR
 
 | |
5N05
 
 | | X-ray crystal structure of an LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.-C.N, Lo Leggio, L. | | Deposit date: | 2017-02-02 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Unliganded and substrate bound structures of the cellooligosaccharide active lytic polysaccharide monooxygenase LsAA9A at low pH.
Carbohydr. Res., 448, 2017
|
|
6S38
 
 | | Ligand binding domain of the P. putida receptor PcaY_PP in complex with quinate | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, Aromatic acid chemoreceptor | | Authors: | Gavira, J.A, Mantilla, M.A, Fernandez, M, Krell, T. | | Deposit date: | 2019-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
5N8H
 
 | | Serial Cu nitrite reductase structures at elevated cryogenic temperature, 240K. Dataset 3. | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, SULFATE ION | | Authors: | Horrell, S, Kekilli, D, Hough, M, Strange, R. | | Deposit date: | 2017-02-23 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Active-site protein dynamics and solvent accessibility in native Achromobacter cycloclastes copper nitrite reductase.
IUCrJ, 4, 2017
|
|
6S3M
 
 | | Crystal structure of helicase Pif1 from Thermus oshimai in complex with ssDNA (dT)18 and ADP-AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*G)-3'), MAGNESIUM ION, ... | | Authors: | Dai, Y.X, Chen, W.F, Teng, F.Y, Liu, N.N, Hou, X.M, Dou, S.X, Rety, S, Xi, X.G. | | Deposit date: | 2019-06-25 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | Structural and functional studies of SF1B Pif1 from Thermus oshimai reveal dimerization-induced helicase inhibition.
Nucleic Acids Res., 49, 2021
|
|
5N8U
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with CTCTCCT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*CP*TP*CP*TP*CP*CP*CP*T)-3'), ... | | Authors: | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural and mechanistic insights into DHX36-mediated innate immunity and G-quadruplex unfolding
To Be Published
|
|
6S40
 
 | | Fragment AZ-001 binding at the p53pT387/14-3-3 sigma interface and additional sites | | Descriptor: | 14-3-3 protein sigma, 4-chloranyl-1-benzothiophene-2-carboximidamide, CALCIUM ION, ... | | Authors: | Leysen, S, Guillory, X, Wolter, M, Genet, S, Somsen, B, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-06-26 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
5N9E
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with TGGGGATTT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*TP*GP*GP*GP*GP*AP*TP*TP*T)-3') | | Authors: | Chen, W.-F, Rety, S, Hai-Lei Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.007 Å) | | Cite: | Structural and mechanistic insights into DHX36-mediated innate immunity and G-quadruplex unfolding
To Be Published
|
|
5NA0
 
 | | TTK kinase domain in complex with a PEG-linked pyrimido-indolizine | | Descriptor: | Dual specificity protein kinase TTK, ~{N}-[3,5-diethyl-1-[2-[2-(2-methoxyethoxy)ethoxy]ethyl]pyrazol-4-yl]-2-[[2-methoxy-4-[4-(2-methoxyethanoyl)piperazin-1-yl]phenyl]amino]-5,6-dihydropyrimido[4,5-e]indolizine-7-carboxamide | | Authors: | Uitdehaag, J, Willemsen-Seegers, N, de Man, J, Buijsman, R.C, Zaman, G.J.R. | | Deposit date: | 2017-02-27 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Target Residence Time-Guided Optimization on TTK Kinase Results in Inhibitors with Potent Anti-Proliferative Activity.
J. Mol. Biol., 429, 2017
|
|
5NA7
 
 | | Structure of DPP III from Bacteroides thetaiotaomicron | | Descriptor: | CHLORIDE ION, Putative dipeptidyl-peptidase III, SULFATE ION, ... | | Authors: | Sabljic, I, Luic, M. | | Deposit date: | 2017-02-27 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Crystal structure of dipeptidyl peptidase III from the human gut symbiont Bacteroides thetaiotaomicron.
PLoS ONE, 12, 2017
|
|
6S26
 
 | | Crystal structure of human wild type STING in complex with 2'-3'-cyclic-GMP-7-deaza-AMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{S},15~{R},17~{R},18~{R})-8-(4-azanylpyrrolo[2,3-d]pyrimidin-7-yl)-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M, Brynda, J. | | Deposit date: | 2019-06-20 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Enzymatic Preparation of 2'-5',3'-5'-Cyclic Dinucleotides, Their Binding Properties to Stimulator of Interferon Genes Adaptor Protein, and Structure/Activity Correlations.
J.Med.Chem., 62, 2019
|
|
5N1W
 
 | | Structure of xEco2 acetyltransferase domain bound to K105-CoA conjugate | | Descriptor: | (2~{S})-2-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethylsulfanyl]propanoic acid, ILE-GLY-ALA-LYX-LYS-ALA,ILE-GLY-ALA-LYX-LYS-ALA,ILE-GLY-ALA-LYX-LYS-ALA,ILE-GLY-ALA-LYX-LYS-ALA, MAGNESIUM ION, ... | | Authors: | Chao, W.C.H, Wade, B.O, Singleton, M.R. | | Deposit date: | 2017-02-06 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Eco1-Mediated Cohesin Acetylation.
Sci Rep, 7, 2017
|
|
6S3O
 
 | | Crystal structure of helicase Pif1 from Thermus oshimai in complex with ssDNA (dT)18 and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Dai, Y.X, Chen, W.F, Teng, F.Y, Liu, N.N, Hou, X.M, Dou, S.X, Rety, S, Xi, X.G. | | Deposit date: | 2019-06-25 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.974 Å) | | Cite: | Structural and functional studies of SF1B Pif1 from Thermus oshimai reveal dimerization-induced helicase inhibition.
Nucleic Acids Res., 49, 2021
|
|
5NBL
 
 | | Crystal structure of the Arp4-N-actin(APO-state) heterodimer bound by a nanobody | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-related protein 4, ... | | Authors: | Knoll, K.R, Eustermann, S, Hopfner, K.P. | | Deposit date: | 2017-03-02 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The nuclear actin-containing Arp8 module is a linker DNA sensor driving INO80 chromatin remodeling.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6S57
 
 | |
5N5G
 
 | |
6S62
 
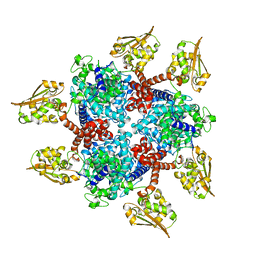 | |
5N7W
 
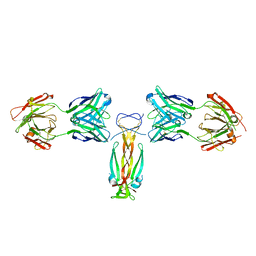 | | Computationally designed functional antibody | | Descriptor: | Antibody Fragment Heavy Chain, Antibody Fragment Light Chain, Interleukin-17A | | Authors: | Hargreaves, D, Breed, J. | | Deposit date: | 2017-02-21 | | Release date: | 2018-11-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Computational Design of Epitope-Specific Functional Antibodies.
Cell Rep, 25, 2018
|
|
5N8G
 
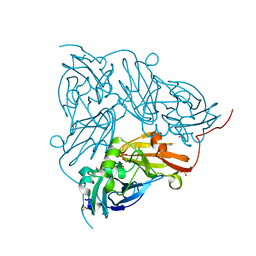 | | Serial Cu nitrite reductase structures at elevated cryogenic temperature, 240K. Dataset 2. | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, SULFATE ION | | Authors: | Horrell, S, Kekilli, D, Hough, M, Strange, R. | | Deposit date: | 2017-02-23 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Active-site protein dynamics and solvent accessibility in native Achromobacter cycloclastes copper nitrite reductase.
IUCrJ, 4, 2017
|
|
6S7C
 
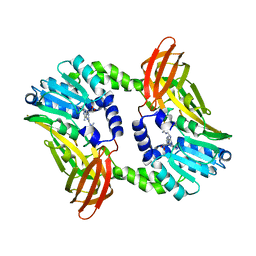 | | Crystal structure of CARM1 in complex with inhibitor UM079 | | Descriptor: | 1-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(3-azanylpropyl)amino]propyl]guanidine, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
5N8Z
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with CTCTCCCTT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*CP*TP*CP*TP*CP*CP*CP*TP*T)-3'), ... | | Authors: | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.477 Å) | | Cite: | Structural and mechanistic insights into DHX36-mediated innate immunity and G-quadruplex unfolding
To Be Published
|
|
