4TVF
 
 | | OxyB from Actinoplanes teichomyceticus | | Descriptor: | OxyB, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Haslinger, K, Cryle, M.J. | | Deposit date: | 2014-06-26 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cytochrome P450 OxyBtei Catalyzes the First Phenolic Coupling Step in Teicoplanin Biosynthesis.
Chembiochem, 15, 2014
|
|
5YRN
 
 | | Structure of RIP2 CARD domain | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Wu, B, Gong, Q. | | Deposit date: | 2017-11-09 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of RIP2 activation and signaling.
Nat Commun, 9, 2018
|
|
5YZ0
 
 | | Cryo-EM Structure of human ATR-ATRIP complex | | Descriptor: | ATR-interacting protein, Serine/threonine-protein kinase ATR | | Authors: | Rao, Q, Liu, M, Tian, Y, Wu, Z, Wang, H, Wang, J, Xu, Y. | | Deposit date: | 2017-12-11 | | Release date: | 2018-01-31 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM structure of human ATR-ATRIP complex.
Cell Res., 28, 2018
|
|
1YSM
 
 | | NMR Structure of N-terminal domain (Residues 1-77) of Siah-Interacting Protein. | | Descriptor: | Calcyclin-binding protein | | Authors: | Bhattacharya, S, Lee, Y.T, Michowski, W, Jastrzebska, B, Filipek, A, Kuznicki, J, Chazin, W.J. | | Deposit date: | 2005-02-08 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Modular Structure of SIP Facilitates Its Role in Stabilizing Multiprotein Assemblies.
Biochemistry, 44, 2005
|
|
1AKP
 
 | | SEQUENTIAL 1H,13C AND 15N NMR ASSIGNMENTS AND SOLUTION CONFORMATION OF APOKEDARCIDIN | | Descriptor: | APOKEDARCIDIN | | Authors: | Constantine, K.L, Colson, K.L, Wittekind, M, Friedrichs, M.S, Zein, N, Tuttle, J, Langley, D.R, Leet, J.E, Schroeder, D.R, Lam, K.S, Farmer II, B.T, Metzler, W.J, Bruccoleri, R.E, Mueller, L. | | Deposit date: | 1994-06-20 | | Release date: | 1994-08-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Sequential 1H, 13C, and 15N NMR assignments and solution conformation of apokedarcidin.
Biochemistry, 33, 1994
|
|
3IIK
 
 | |
4D28
 
 | |
4NQ0
 
 | | Structural insights into yeast histone chaperone Hif1: a scaffold protein recruiting protein complexes to core histones | | Descriptor: | HAT1-interacting factor 1 | | Authors: | Liu, H, Zhang, M, He, W, Zhu, Z, Teng, M, Gao, Y, Niu, L. | | Deposit date: | 2013-11-23 | | Release date: | 2014-07-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into yeast histone chaperone Hif1: a scaffold protein recruiting protein complexes to core histones
Biochem.J., 462, 2014
|
|
3M9V
 
 | |
4A9A
 
 | | Structure of Rbg1 in complex with Tma46 dfrp domain | | Descriptor: | RIBOSOME-INTERACTING GTPASE 1, TRANSLATION MACHINERY-ASSOCIATED PROTEIN 46 | | Authors: | Francis, S.M, Gas, M, Daugeron, M, Seraphin, B, Bravo, J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Rbg1-Tma46 Dimer Structure Reveals New Functional Domains and Their Role in Polysome Recruitment.
Nucleic Acids Res., 40, 2012
|
|
3O1A
 
 | |
6LUF
 
 | |
5KHQ
 
 | | Rasip1 RA domain | | Descriptor: | GLYCEROL, Ras-interacting protein 1 | | Authors: | Gingras, A.R. | | Deposit date: | 2016-06-15 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Dimeric Rasip1 RA Domain Recognition of the Ras Subfamily of GTP-Binding Proteins.
Structure, 24, 2016
|
|
5KHO
 
 | | Rasip1 RA domain in complex with Rap1B | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Gingras, A.R. | | Deposit date: | 2016-06-15 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural Basis of Dimeric Rasip1 RA Domain Recognition of the Ras Subfamily of GTP-Binding Proteins.
Structure, 24, 2016
|
|
3VWI
 
 | | High resolution crystal structure of FraC in the monomeric form | | Descriptor: | AMMONIUM ION, CHLORIDE ION, Fragaceatoxin C, ... | | Authors: | Tanaka, K, Morante, K, Caaveiro, J.M.M, Gonzalez-Manas, J.M, Tsumoto, K. | | Deposit date: | 2012-08-23 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
8TWE
 
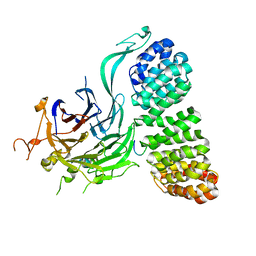 | | Cryo-EM structure of the PP2A:B55-FAM122A complex, B55 body | | Descriptor: | PPP2R1A-PPP2R2A-interacting phosphatase regulator 1, Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Fuller, J.R, Padi, S.K.R, Peti, W, Page, R. | | Deposit date: | 2023-08-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM structures of PP2A:B55-FAM122A and PP2A:B55-ARPP19.
Nature, 625, 2024
|
|
2ZYN
 
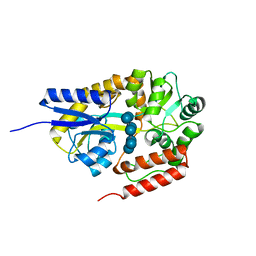 | | Crystal structure of cyclo/maltodextrin-binding protein complexed with beta-cyclodextrin | | Descriptor: | Cycloheptakis-(1-4)-(alpha-D-glucopyranose), Solute-binding protein | | Authors: | Matsumoto, M, Yamada, M, Kurakata, Y, Yoshida, H, Kamitori, S, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2009-01-27 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of open and closed forms of cyclo/maltodextrin-binding protein
Febs J., 276, 2009
|
|
7LY6
 
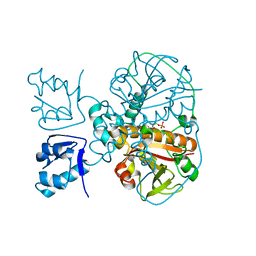 | | Structure of a trans-acting NRPS oxidase, BmdC, involved in bacillamide biosynthesis | | Descriptor: | BmdC, NRPS oxidase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Fortinez, C.M, Bloudoff, K, Schmeing, T.M. | | Deposit date: | 2021-03-05 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structures and function of a tailoring oxidase in complex with a nonribosomal peptide synthetase module.
Nat Commun, 13, 2022
|
|
3W9P
 
 | |
2ZYO
 
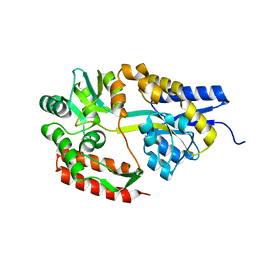 | | Crystal structure of cyclo/maltodextrin-binding protein complexed with maltotetraose | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, solute-binding protein | | Authors: | Matsumoto, M, Yamada, M, Kurakata, Y, Yoshida, H, Kamitori, S, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2009-01-27 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of open and closed forms of cyclo/maltodextrin-binding protein
Febs J., 276, 2009
|
|
2ZYM
 
 | | Crystal structure of cyclo/maltodextrin-binding protein complexed with alpha-cyclodextrin | | Descriptor: | Cyclohexakis-(1-4)-(alpha-D-glucopyranose), Solute-binding protein | | Authors: | Matsumoto, M, Yamada, M, Kurakata, Y, Yoshida, H, Kamitori, S, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2009-01-27 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of open and closed forms of cyclo/maltodextrin-binding protein
Febs J., 276, 2009
|
|
5S8F
 
 | | XChem group deposition -- Crystal Structure of the second bromodomain of pleckstrin homology domain interacting protein (PHIP) in complex with N00572d (space group C2) | | Descriptor: | PH-interacting protein, ~{N}-(5-oxidanylidene-7,8-dihydro-6~{H}-naphthalen-2-yl)ethanamide | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | XChem group deposition
To Be Published
|
|
1W3M
 
 | | Crystal structure of tsushimycin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Delta-3isotetradecenoic acid, ... | | Authors: | Bunkoczi, G, Vertesy, L, Sheldrick, G.M. | | Deposit date: | 2004-07-16 | | Release date: | 2005-07-27 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure of the lipopeptide antibiotic tsushimycin.
Acta Crystallogr. D Biol. Crystallogr., 61, 2005
|
|
5KIU
 
 | | VCP-interacting membrane protein (VIMP) | | Descriptor: | Selenoprotein S | | Authors: | Tang, W.K, Xia, D. | | Deposit date: | 2016-06-17 | | Release date: | 2017-12-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for nucleotide-modulated p97 association with the ER membrane.
Cell Discov, 3, 2017
|
|
5LJ6
 
 | |
