2NRB
 
 | | C28S Mutant of Succinyl-CoA:3-Ketoacid CoA Transferase from Pig Heart | | Descriptor: | CHLORIDE ION, Succinyl-CoA:3-ketoacid-coenzyme A transferase 1 | | Authors: | Tammam, S.D, Fraser, M.E. | | Deposit date: | 2006-11-01 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of the Cysteine Residue Exposed by the Conformational Change in Pig Heart Succinyl-CoA:3-Ketoacid Coenzyme A Transferase on Binding Coenzyme A.
Biochemistry, 46, 2007
|
|
2XVU
 
 | |
5B2F
 
 | | N,N'-diacetylchitobiose deacetylase from Pyrococcus horikoshii complexed with its inhibitor MPG (phosphate-containing condition) | | Descriptor: | 2-deoxy-2-{[(S)-hydroxy(methyl)phosphoryl]amino}-beta-D-glucopyranose, Putative uncharacterized protein PH0499, ZINC ION | | Authors: | Nakamura, T, Niiyama, M, Ida, K, Uegaki, K. | | Deposit date: | 2016-01-15 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate recognition of N,N'-diacetylchitobiose deacetylase from Pyrococcus horikoshii
J.Struct.Biol., 195, 2016
|
|
6T9G
 
 | | CRYSTAL STRUCTURE OF AN ENDOGLUCANASE D213A FROM PENICILLIUM VERRUCULOSUM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoglucanase | | Authors: | Nemashkalov, V, Kravchenko, O, Gabdulkhakov, A, Tischenko, S, Rozhkova, A, Sinitsyn, A. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.29620671 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ENDOGLUCANASE D213A FROM PENICILLIUM VERRUCULOSUM
To Be Published
|
|
2NTS
 
 | | Crystal Structure of SEK-hVb5.1 | | Descriptor: | Staphylococcal enterotoxin K, TRBC1 protein | | Authors: | Gunther, S, Varma, A.K, Moza, B, Sundberg, E.J. | | Deposit date: | 2006-11-08 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel loop domain in superantigens extends their T cell receptor recognition site
J.Mol.Biol., 371, 2007
|
|
6TAF
 
 | | Bd0314 DslA E154Q mutant | | Descriptor: | ACETATE ION, SLT domain-containing protein, SULFATE ION | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2019-10-29 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.336 Å) | | Cite: | A lysozyme with altered substrate specificity facilitates prey cell exit by the periplasmic predator Bdellovibrio bacteriovorus.
Nat Commun, 11, 2020
|
|
2O8C
 
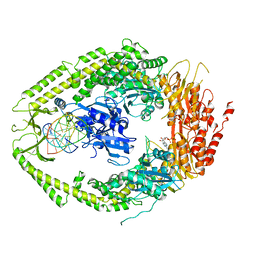 | | human MutSalpha (MSH2/MSH6) bound to ADP and an O6-methyl-guanine T mispair | | Descriptor: | 5'-D(*CP*CP*TP*AP*GP*CP*GP*TP*GP*CP*GP*GP*TP*TP*C)-3', 5'-D(*GP*AP*AP*CP*CP*GP*CP*(6OG)P*CP*GP*CP*TP*AP*GP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Warren, J.J, Pohlhaus, T.J, Changela, A, Modrich, P.L, Beese, L.S. | | Deposit date: | 2006-12-12 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Structure of the Human MutSalpha DNA Lesion Recognition Complex.
Mol.Cell, 26, 2007
|
|
7A77
 
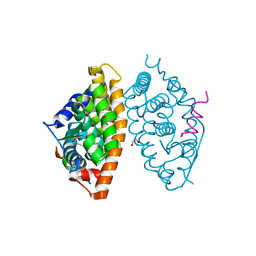 | | Crystal structure of RXR alpha LBD in complexes with palmitic acid and GRIP-1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Nuclear receptor coactivator 2, ... | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comprehensive Set of Tertiary Complex Structures and Palmitic Acid Binding Provide Molecular Insights into Ligand Design for RXR Isoforms.
Int J Mol Sci, 21, 2020
|
|
2OJY
 
 | |
7A79
 
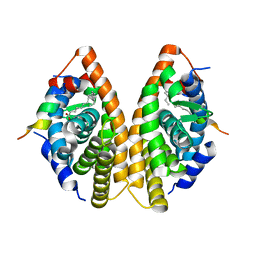 | | Crystal structure of RXR gamma LBD in complexes with palmitic acid and GRIP-1 peptide | | Descriptor: | Nuclear receptor coactivator 2, PALMITIC ACID, Retinoic acid receptor RXR-gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Comprehensive Set of Tertiary Complex Structures and Palmitic Acid Binding Provide Molecular Insights into Ligand Design for RXR Isoforms.
Int J Mol Sci, 21, 2020
|
|
7A7C
 
 | |
5BNT
 
 | |
6TO2
 
 | | Crystal structure of CYP154C5 from Nocardia farcinica in complex with 5alpha-Androstan-3-one | | Descriptor: | (5~{S},8~{S},9~{S},10~{S},13~{S},14~{S})-10,13-dimethyl-1,2,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydrocyclopenta[a]phenanthren-3-one, CHLORIDE ION, Cytochrome P450 monooxygenase, ... | | Authors: | Rodriguez, A, Kluenemann, T, Blankenfeldt, W, Schallmey, A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CYP154C5 Regioselectivity in Steroid Hydroxylation Explored by Substrate Modifications and Protein Engineering*.
Chembiochem, 22, 2021
|
|
2NT0
 
 | | Acid-beta-glucosidase low pH, glycerol bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Glucosylceramidase, ... | | Authors: | Lieberman, R.L, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-11-06 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of acid beta-glucosidase with pharmacological chaperone provides insight into Gaucher disease.
Nat.Chem.Biol., 3, 2007
|
|
6TC9
 
 | | Crystal structure of MutM from Neisseria meningitidis | | Descriptor: | DNA, DNA containing abasic site analogue, Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Silhan, J, Landova, B, Boura, E. | | Deposit date: | 2019-11-05 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.175 Å) | | Cite: | Conformational changes of DNA repair glycosylase MutM triggered by DNA binding.
Febs Lett., 594, 2020
|
|
6HW1
 
 | | ROOM TEMPERATURE STRUCTURE OF LIPASE FROM T. LANUGINOSA AT 2.5 A RESOLUTION IN CHIPX MICROFLUIDIC DEVICE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase, MAGNESIUM ION, ... | | Authors: | Gavira, J.A, Fernadez-Penas, R, Martinez-Rodriguez, S, Verdugo-Escamilla, C. | | Deposit date: | 2018-10-11 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
2K2I
 
 | | NMR Solution structure of the C-terminal domain (T94-Y172) of the human centrin 2 in complex with a repeat sequence of human Sfi1 (R641-T660) | | Descriptor: | Centrin-2, SFI1 peptide | | Authors: | Martinez-Sanz, J, Assairi, L, Blouquit, Y, Duchambon, P, Mouawad, L, Craescu, C. | | Deposit date: | 2008-04-02 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and thermodynamics of the human centrin 2/hSfi1 complex
J.Mol.Biol., 395, 2010
|
|
6RCD
 
 | | Octamer C-Domain P140 Mycoplasma genitalium. | | Descriptor: | MgPa adhesin | | Authors: | Vizarraga, D, Aparicio, D, Perez, R, Illanes, R, Fita, I. | | Deposit date: | 2019-04-11 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Alternative conformation of the C-domain of the P140 protein from Mycoplasma genitalium.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
5AX2
 
 | | Crystal structure of S.cerevisiae Kti11p | | Descriptor: | CADMIUM ION, Diphthamide biosynthesis protein 3 | | Authors: | Kumar, A, Nagarathinam, K, Tanabe, M, Balbach, J. | | Deposit date: | 2015-07-13 | | Release date: | 2016-07-20 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hyperbolic Pressure-Temperature Phase Diagram of the Zinc-Finger Protein apoKti11 Detected by NMR Spectroscopy.
J Phys Chem B, 123, 2019
|
|
2JAE
 
 | | The structure of L-amino acid oxidase from Rhodococcus opacus in the unbound state | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID OXIDASE | | Authors: | Faust, A, Niefind, K, Hummel, W, Schomburg, D. | | Deposit date: | 2006-11-27 | | Release date: | 2007-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Structure of a Bacterial L-Amino Acid Oxidase from Rhodococcus Opacus Gives New Evidence for the Hydride Mechanism for Dehydrogenation.
J.Mol.Biol., 367, 2007
|
|
2XYR
 
 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-18 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
7A62
 
 | | Structure of human indoleamine-2,3-dioxygenase 1 (hIDO1) with a complete JK loop | | Descriptor: | CHLORIDE ION, GLYCEROL, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Mirgaux, M, Wouters, J. | | Deposit date: | 2020-08-24 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.43796444 Å) | | Cite: | Influence of the presence of the heme cofactor on the JK-loop structure in indoleamine 2,3-dioxygenase 1.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
2NVK
 
 | | Crystal Structure of Thioredoxin Reductase from Drosophila melanogaster | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Thioredoxin Reductase | | Authors: | Eckenroth, B.E, Rould, M.A, Hondal, R.J, Everse, S.J. | | Deposit date: | 2006-11-13 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Biochemical Studies Reveal Differences in the Catalytic Mechanisms of Mammalian and Drosophila melanogaster Thioredoxin Reductases.
Biochemistry, 46, 2007
|
|
2NSX
 
 | | Structure of acid-beta-glucosidase with pharmacological chaperone provides insight into Gaucher disease | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, GLYCEROL, ... | | Authors: | Lieberman, R.L, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-11-06 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure of acid beta-glucosidase with pharmacological chaperone provides insight into Gaucher disease.
Nat.Chem.Biol., 3, 2007
|
|
5XXF
 
 | | Crystal structure of Poz1, Tpz1 and Rap1 | | Descriptor: | Protection of telomeres protein poz1, Protection of telomeres protein tpz1, Rap1, ... | | Authors: | Xue, J, Chen, H, Wu, J, Lei, M. | | Deposit date: | 2017-07-03 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the fission yeast S. pombe telomeric Tpz1-Poz1-Rap1 complex.
Cell Res., 27, 2017
|
|
