1XU4
 
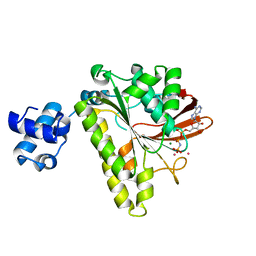 | | ATPASE IN COMPLEX WITH AMP-PNP, MAGNESIUM AND POTASSIUM CO-F | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Wu, Y, Qian, X, He, Y, Moya, I.A, Luo, Y. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an ATPase-active form of Rad51 homolog from Methanococcus voltae. Insights into potassium dependence
J.Biol.Chem., 280, 2005
|
|
1XU5
 
 | | Soluble methane monooxygenase hydroxylase-phenol soaked | | Descriptor: | FE (III) ION, HYDROXIDE ION, Methane monooxygenase component A alpha chain, ... | | Authors: | Sazinsky, M.H, Lippard, S.J. | | Deposit date: | 2004-10-25 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Product Bound Structures of the Soluble Methane Monooxygenase Hydroxylase from Methylococcus capsulatus (Bath): Protein Motion in the Alpha-Subunit
J.Am.Chem.Soc., 127, 2005
|
|
1XU6
 
 | | Structure of the C-terminal domain from Trypanosoma brucei Variant Surface Glycoprotein MITat1.2 | | Descriptor: | Variant surface glycoprotein MITAT 1.2 | | Authors: | Chattopadhyay, A, Jones, N.G, Nietlispach, D, Nielsen, P.R, Voorheis, H.P, Mott, H.R, Carrington, M. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal domain from Trypanosoma brucei variant surface glycoprotein MITat1.2
J.Biol.Chem., 280, 2004
|
|
1XU7
 
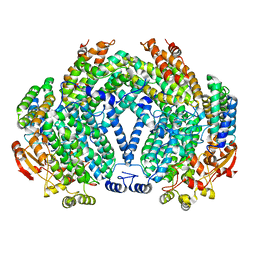
 | | Crystal Structure of the Interface Open Conformation of Tetrameric 11b-HSD1 | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Corticosteroid 11-beta-dehydrogenase, isozyme 1, ... | | Authors: | Hosfield, D.J, Wu, Y, Skene, R.J, Hilger, M, Jennings, A, Snell, G.P, Aertgeerts, K. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational Flexibility in Crystal Structures of Human 11beta-hydroxysteroid dehydrogenase type I provide insights into glucocorticoid interconversion and enzyme regulation.
J.Biol.Chem., 280, 2005
|
|
1XU8
 
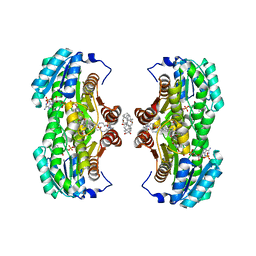
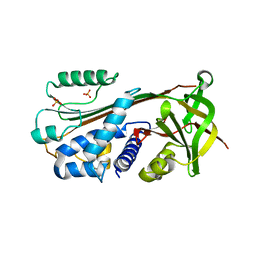 | | The 2.8 A structure of a tumour suppressing serpin | | Descriptor: | Maspin, SULFATE ION | | Authors: | Irving, J.A, Law, R.H, Ruzyla, K, Bashtannyk-Puhalovich, T.A, Kim, N, Worrall, D.M, Rossjohn, J, Whisstock, J.C. | | Deposit date: | 2004-10-25 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The high resolution crystal structure of the human tumor suppressor maspin reveals a novel conformational switch in the G-helix.
J.Biol.Chem., 280, 2005
|
|
1XU9
 
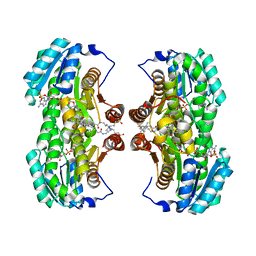 | | Crystal Structure of the Interface Closed Conformation of 11b-hydroxysteroid dehydrogenase isozyme 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Corticosteroid 11-beta-dehydrogenase, ... | | Authors: | Hosfield, D.J, Wu, Y, Skene, R.J, Hilger, M, Jennings, A, Snell, G.P, Aertgeerts, K. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Conformational Flexibility in Crystal Structures of Human 11beta-hydroxysteroid dehydrogenase type I provide insights into glucocorticoid interconversion and enzyme regulation.
J.Biol.Chem., 280, 2005
|
|
1XUA
 
 | | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens | | Descriptor: | (2S,3S)-TRANS-2,3-DIHYDRO-3-HYDROXYANTHRANILIC ACID, Phenazine biosynthesis protein phzF | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1XUB
 
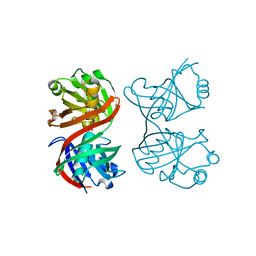 | | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens | | Descriptor: | Phenazine biosynthesis protein phzF, SULFATE ION | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
1XUC
 
 | | Matrix metalloproteinase-13 complexed with non-zinc binding inhibitor | | Descriptor: | CALCIUM ION, Collagenase 3, N,N'-BIS(3-METHYLBENZYL)PYRIMIDINE-4,6-DICARBOXAMIDE, ... | | Authors: | Engel, C.K, Wendt, K.U. | | Deposit date: | 2004-10-26 | | Release date: | 2005-10-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the highly selective inhibition of MMP-13
Chem.Biol., 12, 2005
|
|
1XUD
 
 | | Matrix metalloproteinase-13 complexed with non-zinc binding inhibitor | | Descriptor: | CALCIUM ION, Collagenase 3, N,N'-BIS(4-FLUORO-3-METHYLBENZYL)PYRIMIDINE-4,6-DICARBOXAMIDE, ... | | Authors: | Engel, C.K, Wendt, K.U. | | Deposit date: | 2004-10-26 | | Release date: | 2005-10-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the highly selective inhibition of MMP-13.
Chem.Biol., 12, 2005
|
|
1XUE
 
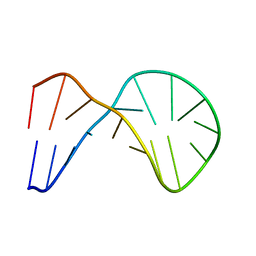 | |
1XUF
 
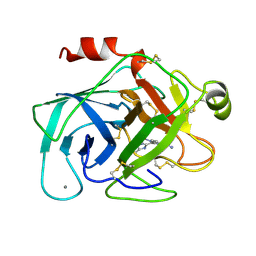 | | TRYPSIN-BABIM-ZN+2, PH 8.2 | | Descriptor: | BIS(5-AMIDINO-BENZIMIDAZOLYL)METHANE ZINC, CALCIUM ION, TRYPSIN | | Authors: | Katz, B.A, Clark, J.M, Finer-Moore, J.S, Jenkins, T.E, Johnson, C.R, Rose, M.J, Luong, C, Moore, W.R, Stroud, R.M. | | Deposit date: | 1997-10-10 | | Release date: | 1998-12-16 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of potent selective zinc-mediated serine protease inhibitors.
Nature, 391, 1998
|
|
1XUG
 
 | | TRYPSIN-BABIM-ZN+2, PH 8.2 | | Descriptor: | BIS(5-AMIDINO-BENZIMIDAZOLYL)METHANE, CALCIUM ION, TRYPSIN, ... | | Authors: | Katz, B.A, Clark, J.M, Finer-Moore, J.S, Jenkins, T.E, Johnson, C.R, Rose, M.J, Luong, C, Moore, W.R, Stroud, R.M. | | Deposit date: | 1997-10-10 | | Release date: | 1998-12-16 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of potent selective zinc-mediated serine protease inhibitors.
Nature, 391, 1998
|
|
1XUH
 
 | | TRYPSIN-KETO-BABIM-CO+2, PH 8.2 | | Descriptor: | BIS(5-AMIDINO-2-BENZIMIDAZOLYL)METHANONE, CALCIUM ION, COBALT (II) ION, ... | | Authors: | Katz, B.A, Clark, J.M, Finer-Moore, J.S, Jenkins, T.E, Johnson, C.R, Rose, M.J, Luong, C, Moore, W.R, Stroud, R.M. | | Deposit date: | 1997-10-10 | | Release date: | 1998-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design of potent selective zinc-mediated serine protease inhibitors.
Nature, 391, 1998
|
|
1XUI
 
 | | TRYPSIN-KETO-BABIM, ZN+2-FREE, PH 8.2 | | Descriptor: | BIS(5-AMIDINO-2-BENZIMIDAZOLYL)METHANONE, CALCIUM ION, SODIUM ION, ... | | Authors: | Katz, B.A, Clark, J.M, Finer-Moore, J.S, Jenkins, T.E, Johnson, C.R, Rose, M.J, Luong, C, Moore, W.R, Stroud, R.M. | | Deposit date: | 1997-10-10 | | Release date: | 1998-11-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of potent selective zinc-mediated serine protease inhibitors.
Nature, 391, 1998
|
|
1XUJ
 
 | | TRYPSIN-KETO-BABIM-ZN+2, PH 8.2 | | Descriptor: | BIS(5-AMIDINO-BENZIMIDAZOLYL)METHANONE ZINC, CALCIUM ION, SULFATE ION, ... | | Authors: | Katz, B.A, Clark, J.M, Finer-Moore, J.S, Jenkins, T.E, Johnson, C.R, Rose, M.J, Luong, C, Moore, W.R, Stroud, R.M. | | Deposit date: | 1997-10-10 | | Release date: | 1998-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Design of potent selective zinc-mediated serine protease inhibitors.
Nature, 391, 1998
|
|
1XUK
 
 | | TRYPSIN-BABIM-SULFATE, PH 5.9 | | Descriptor: | BIS(5-AMIDINO-BENZIMIDAZOLYL)METHANE, CALCIUM ION, SULFATE ION, ... | | Authors: | Katz, B.A, Clark, J.M, Finer-Moore, J.S, Jenkins, T.E, Johnson, C.R, Rose, M.J, Luong, C, Moore, W.R, Stroud, R.M. | | Deposit date: | 1997-10-10 | | Release date: | 1998-11-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design of potent selective zinc-mediated serine protease inhibitors.
Nature, 391, 1998
|
|
1XUO
 
 | | X-ray structure of LFA-1 I-domain bound to a 1,4-diazepane-2,5-dione inhibitor at 1.8A resolution | | Descriptor: | (2R)-2-[3-ISOBUTYL-2,5-DIOXO-4-(QUINOLIN-3-YLMETHYL)-1,4-DIAZEPAN-1-YL]-N-METHYL-3-(2-NAPHTHYL)PROPANAMIDE, Integrin alpha-L, MAGNESIUM ION | | Authors: | Wattanasin, S, Kallen, J, Myers, S, Guo, Q, Sabio, M, Ehrhardt, C, Albert, R, Hommel, U, Weckbecker, G, Welzenbach, K. | | Deposit date: | 2004-10-26 | | Release date: | 2005-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1,4-Diazepane-2,5-diones as novel inhibitors of LFA-1
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1XUP
 
 | | ENTEROCOCCUS CASSELIFLAVUS GLYCEROL KINASE COMPLEXED WITH GLYCEROL | | Descriptor: | GLYCEROL, Glycerol kinase | | Authors: | Yeh, J.I, Charrier, V, Paulo, J, Hou, L, Darbon, E, Hol, W.G.J, Deutscher, J. | | Deposit date: | 2004-10-26 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of Enterococcal Glycerol Kinase in the Absence and Presence of Glycerol: Correlation of Conformation to Substrate Binding and a Mechanism of Activation by Phosphorylation
Biochemistry, 43, 2004
|
|
1XUQ
 
 | | Crystal Structure of SodA-1 (BA4499) from Bacillus anthracis at 1.8A Resolution. | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Boucher, I.W, Levdikov, V.M, Blagova, E.V, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2004-10-26 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of two superoxide dismutases from Bacillus anthracis reveal a novel active centre.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1XUR
 
 | | Matrix metalloproteinase-13 complexed with non-zinc binding inhibitor | | Descriptor: | CALCIUM ION, Collagenase 3, N,N'-BIS(PYRIDIN-3-YLMETHYL)PYRIMIDINE-4,6-DICARBOXAMIDE, ... | | Authors: | Engel, C.K, Wendt, K.U. | | Deposit date: | 2004-10-26 | | Release date: | 2005-10-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the highly selective inhibition of MMP-13.
Chem.Biol., 12, 2005
|
|
1XUT
 
 | | Solution structure of TACI-CRD2 | | Descriptor: | Tumor necrosis factor receptor superfamily member 13B | | Authors: | Hymowitz, S.G, Patel, D.R, Wallweber, H.J, Runyon, S, Yan, M, Yin, J, Shriver, S.K, Gordon, N.C, Pan, B, Skelton, N.J, Kelley, R.F, Starovasnik, M.A. | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-09 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structures of APRIL-receptor complexes: like BCMA, TACI employs only a single cysteine-rich domain for high affinity ligand binding.
J.Biol.Chem., 280, 2005
|
|
1XUU
 
 | | Crystal structure of sialic acid synthase (NeuB) in complex with Mn2+ and Malate from Neisseria meningitidis | | Descriptor: | D-MALATE, MANGANESE (II) ION, polysialic acid capsule biosynthesis protein SiaC | | Authors: | Gunawan, J, Simard, D, Gilbert, M, Lovering, A.L, Wakarchuk, W.W, Tanner, M.E, Strynadka, N.C. | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mechanistic analysis of sialic acid synthase NeuB from Neisseria meningitidis in complex with Mn2+, phosphoenolpyruvate, and N-acetylmannosaminitol.
J.Biol.Chem., 280, 2005
|
|
1XUV
 
 | | X-Ray Crystal Structure of Protein MM0500 from Methanosarcina mazei. Northeast Structural Genomics Consortium Target MaR10. | | Descriptor: | hypothetical protein MM0500 | | Authors: | Forouhar, F, Abashidze, M, Ciano, M, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Hypothetical Protein from Methanosarcina mazei, Northeast Strcutural Genomics Target MaR10
To be Published
|
|
1XUW
 
 | | Structural rationalization of a large difference in RNA affinity despite a small difference in chemistry between two 2'-O-modified nucleic acid analogs | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(NMT)P*AP*CP*GP*C)-3') | | Authors: | Pattanayek, R, Sethaphong, L, Pan, C, Prhavc, M, Prakash, T.P, Manoharan, M, Egli, M. | | Deposit date: | 2004-10-26 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural rationalization of a large difference in RNA affinity despite a small difference in chemistry between two 2'-O-modified nucleic acid analogues.
J.Am.Chem.Soc., 126, 2004
|
|
