6GS2
 
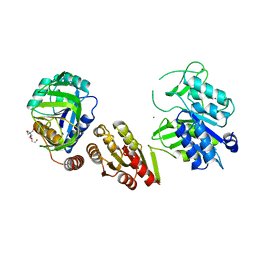 | | Crystal Structure of the GatD/MurT Enzyme Complex from Staphylococcus aureus | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, SA1707 protein, ... | | Authors: | Muckenfuss, L.M, Noeldeke, E.R, Niemann, V, Zocher, G, Stehle, T. | | Deposit date: | 2018-06-13 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural basis of cell wall peptidoglycan amidation by the GatD/MurT complex of Staphylococcus aureus.
Sci Rep, 8, 2018
|
|
2VTT
 
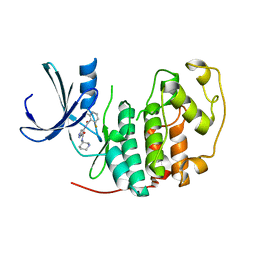 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | 4-{[(2,6-difluorophenyl)carbonyl]amino}-N-[(3S)-piperidin-3-yl]-1H-pyrazole-3-carboxamide, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
4QH3
 
 | |
4AMZ
 
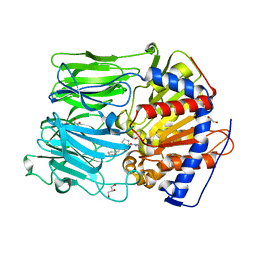 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN WITH A COVALENTLY BOUND INHIBITOR IC-2 | | Descriptor: | (5R)-N-benzyl-5-({(2S)-2-[(1R)-1,2-dihydroxyethyl]pyrrolidin-1-yl}carbonyl)cyclopent-1-ene-1-carboxamide, GLYCEROL, PROLYL ENDOPEPTIDASE | | Authors: | Kaszuba, K, Rog, T, Danne, R, Canning, P, Fulop, V, Juhasz, T, Szeltner, Z, St-Pierre, J.F, Garcia-Horsman, A, Mannisto, P.T, Karttunen, M, Hokkanen, J, Bunker, A. | | Deposit date: | 2012-03-14 | | Release date: | 2012-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Dynamics, Crystallography and Mutagenesis Studies on the Substrate Gating Mechanism of Prolyl Oligopeptidase.
Biochimie, 94, 2012
|
|
2VTH
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H-pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design | | Descriptor: | 5-hydroxynaphthalene-1-sulfonamide, CELL DIVISION PROTEIN KINASE 2, GLYCEROL | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2VTP
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, {[(2,6-difluorophenyl)carbonyl]amino}-N-(4-fluorophenyl)-1H-pyrazole-3-carboxamide | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
4O72
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with NU7441 | | Descriptor: | 1,2-ETHANEDIOL, 8-(dibenzo[b,d]thiophen-4-yl)-2-(morpholin-4-yl)-4H-chromen-4-one, Bromodomain-containing protein 4, ... | | Authors: | Zhu, J.-Y, Ember, S.W, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
1NME
 
 | | Structure of Casp-3 with tethered salicylate | | Descriptor: | 2-HYDROXY-5-(2-MERCAPTO-ETHYLSULFAMOYL)-BENZOIC ACID, 3-(2-MERCAPTO-ACETYLAMINO)-4-OXO-PENTANOIC ACID, Caspase-3 | | Authors: | Erlanson, D.A, Lam, J, Wiesmann, C, Luong, T.N, Simmons, B, DeLano, W, Choong, I.C, Flanagan, M, Lee, D, O'Brian, T. | | Deposit date: | 2003-01-09 | | Release date: | 2003-03-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In situ assembly of enzyme inhibitors using extended tethering.
Nat.Biotechnol., 21, 2003
|
|
1B0W
 
 | |
8F6N
 
 | | Dihydropyrimidine Dehydrogenase (DPD) C671S Mutant Soaked with Thymine Quasi-Anaerobically | | Descriptor: | Dihydropyrimidine dehydrogenase [NADP(+)], FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kaley, N, Smith, M, Forouzesh, D, Liu, D, Moran, G. | | Deposit date: | 2022-11-16 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Mammalian dihydropyrimidine dehydrogenase: Added mechanistic details from transient-state analysis of charge transfer complexes.
Arch.Biochem.Biophys., 736, 2023
|
|
8F5W
 
 | | Dihydropyrimidine Dehydrogenase (DPD) C671S Mutant Soaked with Dihydrothymine and NADPH Quasi-Anaerobically | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, Dihydropyrimidine dehydrogenase [NADP(+)], FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kaley, N, Smith, M, Forouzesh, D, Liu, D, Moran, G. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-01 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mammalian dihydropyrimidine dehydrogenase: Added mechanistic details from transient-state analysis of charge transfer complexes.
Arch.Biochem.Biophys., 736, 2023
|
|
6R35
 
 | | Structure of the LecB lectin from Pseudomonas aeruginosa strain PAO1 in complex with lewis x tetrasaccharide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Fucose-binding lectin PA-IIL, ... | | Authors: | Lepsik, M, Sommer, R, Kuhaudomlarp, S, Lelimousin, M, Varrot, A, Titz, A, Imberty, A. | | Deposit date: | 2019-03-19 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Induction of rare conformation of oligosaccharide by binding to calcium-dependent bacterial lectin: X-ray crystallography and modelling study.
Eur.J.Med.Chem., 177, 2019
|
|
2OHX
 
 | |
6Q4C
 
 | | CDK2 in complex with FragLite16 | | Descriptor: | 4-bromanyl-1,8-naphthyridine, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
8EXG
 
 | | Human Carbonic Anhydrase II bound N-(2-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethyl)-4-sulfamoylbenzamide | | Descriptor: | (4S)-5-amino-4-{1,3-dioxo-4-[(2-{2-[2-(4-sulfamoylbenzamido)ethoxy]ethoxy}ethyl)amino]-1,3-dihydro-2H-isoindol-2-yl}-5-oxopentanoic acid, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Kohlbrand, A.J, O'Herin, C.B. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Development of Human Carbonic Anhydrase II Heterobifunctional Degraders.
J.Med.Chem., 2023
|
|
8F61
 
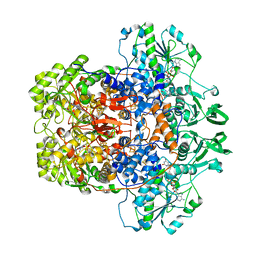 | | Dihydropyrimidine Dehydrogenase (DPD) C671S Mutant Soaked with Dihydrothymine Quasi-Anaerobically | | Descriptor: | (5S)-5-methyl-1,3-diazinane-2,4-dione, 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, Dihydropyrimidine dehydrogenase [NADP(+)], ... | | Authors: | Kaley, N, Smith, M, Forouzesh, D, Liu, D, Moran, G. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Mammalian dihydropyrimidine dehydrogenase: Added mechanistic details from transient-state analysis of charge transfer complexes.
Arch.Biochem.Biophys., 736, 2023
|
|
1BCJ
 
 | |
8EXC
 
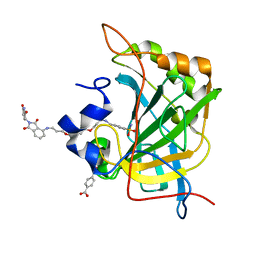 | | Human Carbonic Anhydrase II bound tert-butyl (3-(4-(3-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)propoxy)butoxy)propyl)carbamate | | Descriptor: | Carbonic anhydrase 2, MERCURIBENZOIC ACID, N-(3-{4-[3-({2-[(3R)-2,6-dioxopiperidin-3-yl]-1,3-dioxo-2,3-dihydro-1H-isoindol-4-yl}amino)propoxy]butoxy}propyl)-4-sulfamoylbenzamide, ... | | Authors: | Kohlbrand, A.J, O'Herin, C.B. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of Human Carbonic Anhydrase II Heterobifunctional Degraders.
J.Med.Chem., 2023
|
|
4MHF
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_3107), target EFI-510173, with bound alpha/beta D-Glucuronate, space group P21 | | Descriptor: | TRAP dicarboxylate transporter, DctP subunit, alpha-D-glucopyranuronic acid, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-29 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
8F17
 
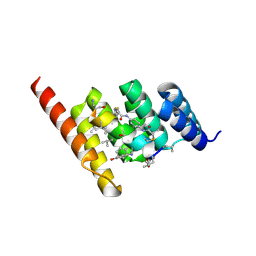 | | Structure of the STUB1 TPR domain in complex with H204, an all-D Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CHIP, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Callahan, A.J, Travaline, T.L, Tokareva, O.S, Swiecicki, J.-M, Verdine, G.L, Pentelute, B.L, McGee, J.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Single-Shot Flow Synthesis of D-Proteins for Mirror-Image Phage Display
Chemrxiv, 2023
|
|
8F15
 
 | | Structure of the STUB1 TPR domain in complex with H202, an all-D Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CHIP, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Callahan, A.J, Travaline, T.L, Tokareva, O.S, Swiecicki, J.-M, Verdine, G.L, Pentelute, B.L, McGee, J.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Single-Shot Flow Synthesis of D-Proteins for Mirror-Image Phage Display
Chemrxiv, 2023
|
|
8EYL
 
 | | Human Carbonic Anhydrase II with Tert-butyl (2-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethyl)carbamate | | Descriptor: | 2-[[(2~{R})-1-azanyl-5-oxidanyl-1,5-bis(oxidanylidene)pentan-2-yl]carbamoyl]-6-[2-[2-[(4-sulfamoylphenyl)carbonylamino]ethoxy]ethylamino]benzoic acid, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Kohlbrand, A.J, O'Herin, C.B. | | Deposit date: | 2022-10-27 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Development of Human Carbonic Anhydrase II Heterobifunctional Degraders.
J.Med.Chem., 2023
|
|
8F16
 
 | | Structure of the STUB1 TPR domain in complex with H203, an all-D Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CHIP, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Callahan, A.J, Travaline, T.L, Tokareva, O.S, Swiecicki, J.-M, Verdine, G.L, Pentelute, B.L, McGee, J.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Single-Shot Flow Synthesis of D-Proteins for Mirror-Image Phage Display
Chemrxiv, 2023
|
|
8F14
 
 | | Structure of the STUB1 TPR domain in complex with H201, an all-D Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CHIP, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Callahan, A.J, Travaline, T.L, Tokareva, O.S, Swiecicki, J.-M, Verdine, G.L, Pentelute, B.L, McGee, J.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Single-Shot Flow Synthesis of D-Proteins for Mirror-Image Phage Display
Chemrxiv, 2023
|
|
6HE1
 
 | | Pseudomonas aeruginosa Seryl-tRNA Synthetase in Complex with 5'-O-(N-(L-seryl)-sulfamoyl)N3-methyluridine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-(N-(L-seryl)-sulfamoyl)N3-methyluridine, SODIUM ION, ... | | Authors: | Pang, L, De Graef, S, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2018-08-20 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural Insights into the Binding of Natural Pyrimidine-Based Inhibitors of Class II Aminoacyl-tRNA Synthetases.
Acs Chem.Biol., 15, 2020
|
|
