5LND
 
 | | Crystal structure of self-complemented MyfA, the major subunit of Myf fimbriae from Yersinia enterocolitica | | Descriptor: | Fimbrial protein MyfA,Fimbrial protein MyfA,Fimbrial protein MyfA | | Authors: | Pakharukova, N.A, Roy, S, Tuitilla, M, Zavialov, A.V. | | Deposit date: | 2016-08-04 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural basis for Myf and Psa fimbriae-mediated tropism of pathogenic strains of Yersinia for host tissues.
Mol.Microbiol., 102, 2016
|
|
5LN4
 
 | | Crystal structure of self-complemented PsaA, the major subunit of pH 6 antigen from Yersinia pests, in complex with choline | | Descriptor: | CHOLINE ION, pH 6 antigen,pH 6 antigen | | Authors: | Pakharukova, N.A, Roy, S, Rahman, M.M, Tuitilla, M, Zavialov, A.V. | | Deposit date: | 2016-08-03 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural basis for Myf and Psa fimbriae-mediated tropism of pathogenic strains of Yersinia for host tissues.
Mol.Microbiol., 102, 2016
|
|
5LO7
 
 | | Crystal structure of self-complemented MyfA, the major subunit of Myf fimbriae from Yersinia enterocolitica | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Fimbrial protein MyfA,Fimbrial protein MyfA | | Authors: | Pakharukova, N.A, Roy, S, Tuitilla, M, Zavialov, A.V. | | Deposit date: | 2016-08-08 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for Myf and Psa fimbriae-mediated tropism of pathogenic strains of Yersinia for host tissues.
Mol.Microbiol., 102, 2016
|
|
7ER3
 
 | | Crystal structure of beta-lactoglobulin complexed with chloroquine | | Descriptor: | (4S)-N~4~-(7-chloroquinolin-4-yl)-N~1~,N~1~-diethylpentane-1,4-diamine, Major allergen beta-lactoglobulin | | Authors: | Yao, Q, Ma, J, Xing, Y, Zang, J. | | Deposit date: | 2021-05-05 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Binding of Chloroquine to Whey Protein Relieves Its Cytotoxicity while Enhancing Its Uptake by Cells.
J.Agric.Food Chem., 69, 2021
|
|
3J41
 
 | | Pseudo-atomic model of the Aquaporin-0/Calmodulin complex derived from electron microscopy | | Descriptor: | CALCIUM ION, Calmodulin, Lens fiber major intrinsic protein | | Authors: | Reichow, S.L, Clemens, D.M, Freites, J.A, Nemeth-Cahalan, K.L, Heyden, M, Tobias, D.J, Hall, J.E, Gonen, T. | | Deposit date: | 2013-05-31 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Allosteric mechanism of water-channel gating by Ca(2+)-calmodulin.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4IU9
 
 | | Crystal structure of a membrane transporter | | Descriptor: | Nitrite extrusion protein 2 | | Authors: | Yan, H, Huang, W, Yan, C, Gong, X, Jiang, S, Zhao, Y, Wang, J, Shi, Y. | | Deposit date: | 2013-01-20 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structure and mechanism of a nitrate transporter.
Cell Rep, 3, 2013
|
|
4IU8
 
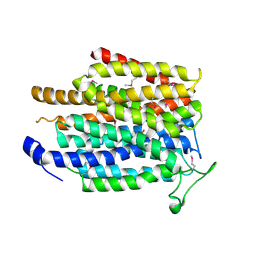 | | Crystal structure of a membrane transporter (selenomethionine derivative) | | Descriptor: | NITRATE ION, Nitrite extrusion protein 2 | | Authors: | Yan, H, Huang, W, Yan, C, Gong, X, Jiang, S, Zhao, Y, Wang, J, Shi, Y. | | Deposit date: | 2013-01-20 | | Release date: | 2013-04-17 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structure and mechanism of a nitrate transporter.
Cell Rep, 3, 2013
|
|
4LDS
 
 | |
5BUP
 
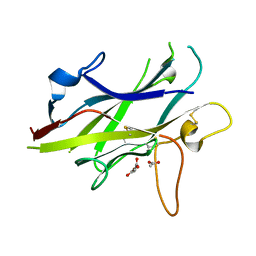 | | Crystal structure of the ZP-C domain of mouse ZP2 | | Descriptor: | ACETATE ION, Zona pellucida sperm-binding protein 2 | | Authors: | Nishimura, K, Jovine, L. | | Deposit date: | 2015-06-04 | | Release date: | 2016-01-27 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | A structured interdomain linker directs self-polymerization of human uromodulin.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1RB8
 
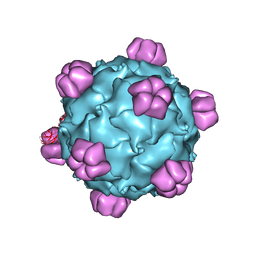 | | The phiX174 DNA binding protein J in two different capsid environments. | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, Capsid protein, DNA (5'-D(P*CP*AP*AP*A)-3'), ... | | Authors: | Bernal, R.A, Hafenstein, S, Esmeralda, R, Fane, B.A, Rossmann, M.G. | | Deposit date: | 2003-11-03 | | Release date: | 2004-04-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The phiX174 Protein J Mediates DNA Packaging and Viral Attachment to Host Cells.
J.Mol.Biol., 337, 2004
|
|
1M06
 
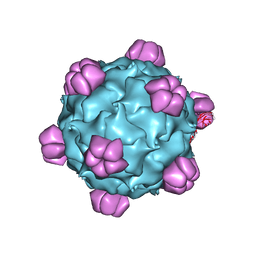 | | Structural Studies of Bacteriophage alpha3 Assembly, X-Ray Crystallography | | Descriptor: | 5'-D(P*(3DR)P*(3DR)P*(3DR)P*(3DR)P*(3DR)P*(3DR)P*(3DR)P*(3DR)P*(3DR)P*(3DR))-3', Capsid Protein, Major spike protein, ... | | Authors: | Bernal, R.A, Hafenstein, S, Olson, N.H, Bowman, V, Chipman, P.R, Baker, T.S, Fane, B.A, Rossmann, M.G. | | Deposit date: | 2002-06-12 | | Release date: | 2002-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Studies of Bacteriophage alpha3 Assembly
J.Mol.Biol., 325, 2003
|
|
1M0F
 
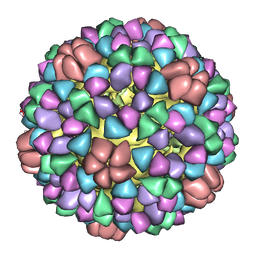 | | Structural Studies of Bacteriophage alpha3 Assembly, Cryo-electron microscopy | | Descriptor: | Capsid Protein F, Major Spike Protein G, Scaffolding Protein B, ... | | Authors: | Bernal, R.A, Hafenstein, S, Olson, N.H, Bowman, V.D, Chipman, P.R, Baker, T.S, Fane, B.A, Rossmann, M.G. | | Deposit date: | 2002-06-12 | | Release date: | 2002-12-25 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Structural Studies of Bacteriophage alpha3 Assembly
J.Mol.Biol., 325, 2003
|
|
6EI3
 
 | | Crystal structure of auto inhibited POT family peptide transporter | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Proton-dependent oligopeptide transporter family protein | | Authors: | Newstead, S, Brinth, A, Vogeley, L, Caffrey, M. | | Deposit date: | 2017-09-17 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Proton movement and coupling in the POT family of peptide transporters.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2JU8
 
 | | Solution-State Structures of Oleate-Liganded LFABP, Major Form of 1:2 Protein-Ligand Complex | | Descriptor: | Fatty acid-binding protein, liver, OLEIC ACID | | Authors: | He, Y, Yang, X, Wang, H, Estephan, R, Francis, F, Kodukula, S, Storch, J, Stark, R.E. | | Deposit date: | 2007-08-15 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution-State Molecular Structure of Apo and Oleate-Liganded Liver Fatty Acid-Binding Protein
Biochemistry, 46, 2007
|
|
7L16
 
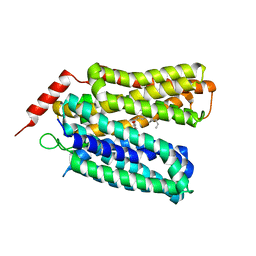 | | Crystal structure of sugar-bound melibiose permease MelB | | Descriptor: | Melibiose carrier protein, dodecyl 6-O-alpha-D-galactopyranosyl-beta-D-glucopyranoside | | Authors: | Guan, L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | X-ray crystallography reveals molecular recognition mechanism for sugar binding in a melibiose transporter MelB.
Commun Biol, 4, 2021
|
|
7L17
 
 | | Crystal structure of sugar-bound melibiose permease MelB | | Descriptor: | 4-nitrophenyl alpha-D-galactopyranoside, Melibiose carrier protein | | Authors: | Guan, L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | X-ray crystallography reveals molecular recognition mechanism for sugar binding in a melibiose transporter MelB.
Commun Biol, 4, 2021
|
|
2X4R
 
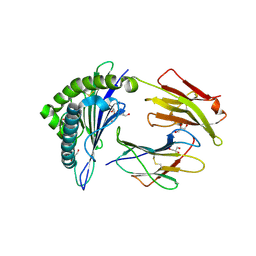 | | Crystal structure of MHC CLass I HLA-A2.1 bound to Cytomegalovirus (CMV) pp65 epitope | | Descriptor: | 65 KDA PHOSPHOPROTEIN, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
2X4O
 
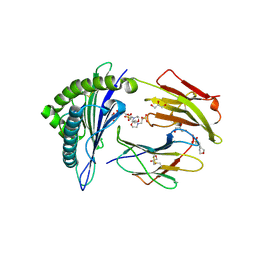 | | Crystal structure of MHC CLass I HLA-A2.1 bound to HIV-1 envelope peptide env120-128 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, ENVELOPE GLYCOPROTEIN GP160, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
2X4N
 
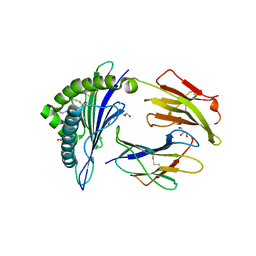 | | Crystal structure of MHC CLass I HLA-A2.1 bound to residual fragments of a photocleavable peptide that is cleaved upon UV-light treatment | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
2X4S
 
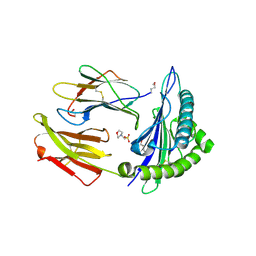 | | Crystal structure of MHC CLass I HLA-A2.1 bound to a peptide representing the epitope of the H5N1 (Avian Flu) Nucleoprotein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals
J.Am.Chem.Soc., 131, 2009
|
|
2X4U
 
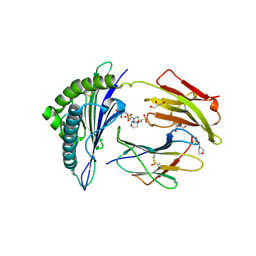 | | Crystal structure of MHC CLass I HLA-A2.1 bound to HIV-1 Peptide RT468-476 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
2X70
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to a photocleavable peptide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-22 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
2CIK
 
 | | Insights Into Crossreactivity in Human Allorecognition: The Structure of HLA-B35011 Presenting an Epitope derived from Cytochrome P450. | | Descriptor: | BETA-2-MICROGLOBULIN, GLYCEROL, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN B-35 ALPHA CHAIN, ... | | Authors: | Hourigan, C.S, Harkiolaki, M, Peterson, N.A, Bell, J.I, Jones, E.Y, O'Callaghan, C.A. | | Deposit date: | 2006-03-22 | | Release date: | 2006-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Structure of the Human Allo-Ligand Hla-B3501 in Complex with a Cytochrome P450 Peptide: Steric Hindrance Influences Tcr Allo-Recognition.
Eur.J.Immunol., 36, 2006
|
|
5VME
 
 | | Crystal structure of human IgG1 Fc K248E, T437R mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, human IgG1 Fc K248E,T437R mutant | | Authors: | Armstrong, A.A, Gilliland, G.L. | | Deposit date: | 2017-04-27 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Functional optimization of agonistic antibodies to OX40 receptor with novel Fc mutations to promote antibody multimerization.
MAbs, 9, 2017
|
|
6J2S
 
 | | Structure of HitA bound to gallium from Pseudomonas aeruginosa | | Descriptor: | Ferric iron-binding periplasmic protein, GALLIUM (III) ION, PHOSPHATE ION | | Authors: | Guo, Y, Li, H.Y, Sun, H.Z, Xia, W. | | Deposit date: | 2019-01-02 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.73005116 Å) | | Cite: | Identification and Characterization of a Metalloprotein Involved in Gallium Internalization in Pseudomonas aeruginosa.
Acs Infect Dis., 5, 2019
|
|
