8TZQ
 
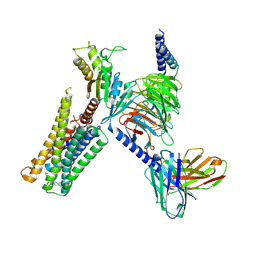 | | CryoEM structure of D2 dopamine receptor in complex with GoA KE Mutant, scFv16, and dopamine | | Descriptor: | D(2) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Krumm, B.E, Kapolka, N.J, Fay, J.F, Roth, B.L. | | Deposit date: | 2023-08-27 | | Release date: | 2024-08-21 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A neurodevelopmental disorder mutation locks G proteins in the transitory pre-activated state.
Nat Commun, 15, 2024
|
|
8U02
 
 | | CryoEM structure of D2 dopamine receptor in complex with GoA KE mutant and dopamine | | Descriptor: | D(2) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Krumm, B.E, Kapolka, N.J, Fay, J.F, Roth, B.L. | | Deposit date: | 2023-08-28 | | Release date: | 2024-08-21 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | A neurodevelopmental disorder mutation locks G proteins in the transitory pre-activated state.
Nat Commun, 15, 2024
|
|
8PNT
 
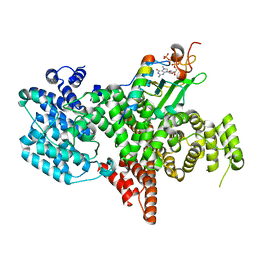 | | Structure of the human nuclear cap-binding complex bound to PHAX and m7G-capped RNA | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Nuclear cap-binding protein subunit 1, Nuclear cap-binding protein subunit 2, ... | | Authors: | Dubiez, E, Pellegrini, E, Foucher, A.E, Cusack, S, Kadlec, J. | | Deposit date: | 2023-07-01 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis for competitive binding of productive and degradative co-transcriptional effectors to the nuclear cap-binding complex.
Cell Rep, 43, 2024
|
|
8UF4
 
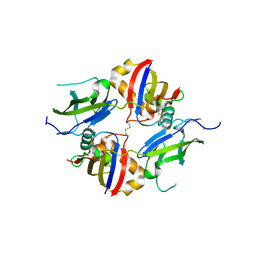 | | Crystal structure of wildtype dystroglycan proteolytic domain (juxtamembrane domain) | | Descriptor: | Beta-dystroglycan, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Anderson, M.J.M, Shi, K, Hayward, A.N, Uhlens, C, Evans III, R.L, Grant, E, Greenberg, L, Aihara, H, Gordon, W.R. | | Deposit date: | 2023-10-03 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Molecular basis of proteolytic cleavage regulation by the extracellular matrix receptor dystroglycan.
Structure, 32, 2024
|
|
7NWG
 
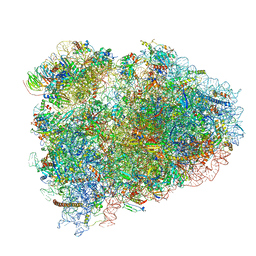 | | Mammalian pre-termination 80S ribosome with Hybrid P/E- and A/P-site tRNA's bound by Blasticidin S. | | Descriptor: | 18S Ribosomal RNA, 28S Ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Powers, K.T, Yadav, S.K.N, Bufton, J.C, Schaffitzel, C. | | Deposit date: | 2021-03-16 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Blasticidin S inhibits mammalian translation and enhances production of protein encoded by nonsense mRNA.
Nucleic Acids Res., 49, 2021
|
|
7PUJ
 
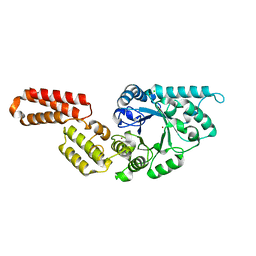 | | Crystal structure of Endoglycosidase E GH18 domain from Enterococcus faecalis | | Descriptor: | Beta-N-acetylhexosaminidase, CHLORIDE ION, ZINC ION | | Authors: | Garcia-Alija, M, Du, J.J, Trastoy, B, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2021-09-30 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Mechanism of cooperative N-glycan processing by the multi-modular endoglycosidase EndoE.
Nat Commun, 13, 2022
|
|
7NL0
 
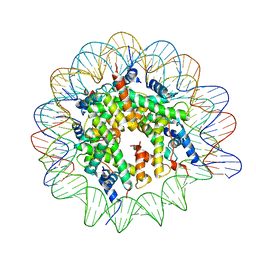 | | Cryo-EM structure of the Lin28B nucleosome core particle | | Descriptor: | DNA (131-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Roberts, G.A, Ozkan, B, Gachulincova, I, O Dwyer, M.R, Hall-Ponsele, E, Saxena, M, Robinson, P.J, Soufi, A. | | Deposit date: | 2021-02-19 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Dissecting OCT4 defines the role of nucleosome binding in pluripotency.
Nat.Cell Biol., 23, 2021
|
|
8UGM
 
 | |
8UIX
 
 | |
8B5Q
 
 | |
8B62
 
 | | Crystal Structure of P. aeruginosa WaaG in complex with UDP-galactose | | Descriptor: | GALACTOSE-URIDINE-5'-DIPHOSPHATE, GLYCEROL, UDP-glucose:(Heptosyl) LPS alpha 1,3-glucosyltransferase WaaG | | Authors: | Scaletti, E, Gustafsson Westergren, R, Stenmark, P. | | Deposit date: | 2022-09-25 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural and functional insights into the Pseudomonas aeruginosa glycosyltransferase WaaG and the implications for lipopolysaccharide biosynthesis.
J.Biol.Chem., 299, 2023
|
|
8B63
 
 | | Crystal Structure of P. aeruginosa WaaG in complex with UDP-GalNAc | | Descriptor: | ACETATE ION, UDP-glucose:(Heptosyl) LPS alpha 1,3-glucosyltransferase WaaG, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Scaletti, E, Gustafsson Westergren, R, Stenmark, P. | | Deposit date: | 2022-09-25 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insights into the Pseudomonas aeruginosa glycosyltransferase WaaG and the implications for lipopolysaccharide biosynthesis.
J.Biol.Chem., 299, 2023
|
|
8B5S
 
 | | Crystal Structure of P. aeruginosa WaaG in complex with UDP-glucose | | Descriptor: | UDP-glucose:(Heptosyl) LPS alpha 1,3-glucosyltransferase WaaG, URIDINE-5'-DIPHOSPHATE, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Scaletti, E, Gustafsson Westergren, R, Stenmark, P. | | Deposit date: | 2022-09-24 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and functional insights into the Pseudomonas aeruginosa glycosyltransferase WaaG and the implications for lipopolysaccharide biosynthesis.
J.Biol.Chem., 299, 2023
|
|
8VAQ
 
 | |
8VAP
 
 | |
8VAM
 
 | |
7NLC
 
 | | Crystallographic structure of human Tsg101 UEV domain in complex with a HEV ORF3 peptide | | Descriptor: | AMMONIUM ION, CHLORIDE ION, Protein ORF3, ... | | Authors: | Moschidi, D, Dupre, E, Villeret, V, Hanoulle, X. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Crystallographic structure of human Tsg101 UEV domain in complex with a HEV ORF3 peptide
To Be Published
|
|
8VAS
 
 | |
8BSP
 
 | | Notum Inhibitor ARUK3006560 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-26 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8BSR
 
 | | Notum Inhibitor ARUK3006562 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-26 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8BT2
 
 | | Notum Inhibitor ARUK3004876 | | Descriptor: | 1,2-ETHANEDIOL, 1-[5-chloranyl-4-(trifluoromethyl)-2,3-dihydroindol-1-yl]ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-27 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8BT7
 
 | | Notum Inhibitor ARUK3004903 | | Descriptor: | 1,2-ETHANEDIOL, 1-[3,4-bis(chloranyl)-5-methyl-indol-1-yl]ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8BT5
 
 | | Notum Inhibitor ARUK3004877 | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-fluoranylspiro[2~{H}-indole-3,1'-cyclobutane]-1-yl)ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-27 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8BSQ
 
 | | Notum Inhibitor ARUK3006561 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-26 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8BTE
 
 | | Notum Inhibitor ARUK3004470 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
