8GEY
 
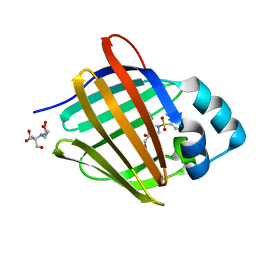 | | Crystal structure of human cellular retinol binding protein 1 in complex with 4-(hydroxymethyl)-1-[(4-methoxy-5,6,7,8-tetrahydronaphthalen-1-yl)sulfonyl]piperidin-4-ol | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(hydroxymethyl)-1-(4-methoxy-5,6,7,8-tetrahydronaphthalene-1-sulfonyl)piperidin-4-ol, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
8GD2
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with N-methyl-1-{3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}-N-(2-thienylmethyl)methanamine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-methyl-1-{3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}-N-[(thiophen-2-yl)methyl]methanamine, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
6JMK
 
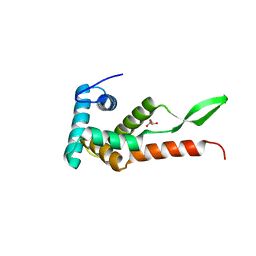 | | Ribosomal protein S7 from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, 30S ribosomal protein S7, GLYCEROL | | Authors: | Li, Z, Li, J. | | Deposit date: | 2019-03-11 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the complex of trigger factor chaperone and ribosomal protein S7 from Mycobacterium tuberculosis.
Biochem. Biophys. Res. Commun., 512, 2019
|
|
4Y3F
 
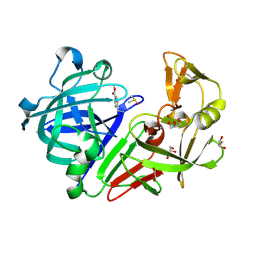 | |
5L67
 
 | |
2L8C
 
 | |
2G48
 
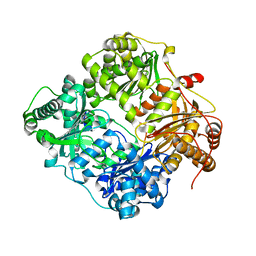 | |
3AHJ
 
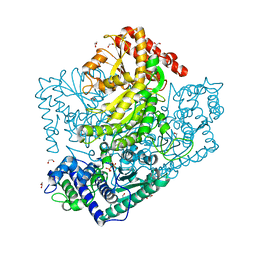 | | H553A mutant of Phosphoketolase from Bifidobacterium Breve | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, THIAMINE DIPHOSPHATE, ... | | Authors: | Suzuki, R, Katayama, T, Kim, B.-J, Wakagi, T, Shoun, H, Ashida, H, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism
J.Biol.Chem., 285, 2010
|
|
6TN1
 
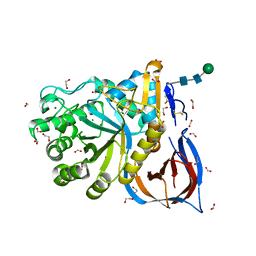 | | Unliganded Crystal Structure of Recombinant GBA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FORMIC ACID, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2019-12-05 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | A baculoviral system for the production of human beta-glucocerebrosidase enables atomic resolution analysis.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
3W06
 
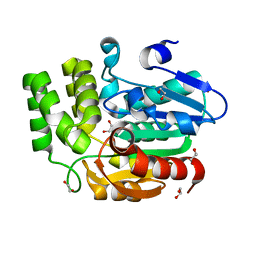 | | Crystal structure of Arabidopsis thaliana DWARF14 Like (AtD14L) | | Descriptor: | 1,2-ETHANEDIOL, Hydrolase, alpha/beta fold family protein | | Authors: | Kagiyama, M, Hirano, Y, Mori, T, Kim, S.Y, Kyozuka, J, Seto, Y, Yamaguchi, S, Hakoshima, T. | | Deposit date: | 2012-10-19 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structures of D14 and D14L in the strigolactone and karrikin signaling pathways
Genes Cells, 18, 2013
|
|
5L5S
 
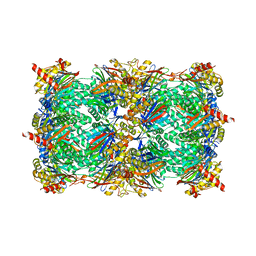 | |
5L6A
 
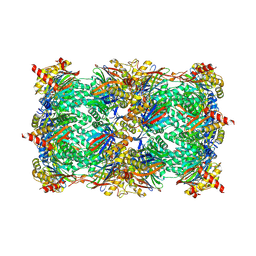 | | Yeast 20S proteasome with mouse beta5i (1-138) and mouse beta6 (97-111; 118-133) in complex with epoxyketone inhibitor 17 | | Descriptor: | (2~{S})-3-(4-methoxyphenyl)-~{N}-[(2~{S},3~{S},4~{R})-4-methyl-3,5-bis(oxidanyl)-1-phenyl-pentan-2-yl]-2-[[(2~{R})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
6GIU
 
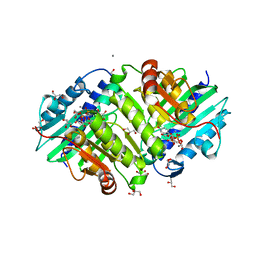 | | Human IMPase with L-690330 | | Descriptor: | GLYCEROL, Inositol monophosphatase 1, MANGANESE (II) ION, ... | | Authors: | Kraft, L.V, Roe, S.M. | | Deposit date: | 2018-05-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Co-crystallization of human inositol monophosphatase with the lithium mimetic L-690,330.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
1WI7
 
 | |
6JV0
 
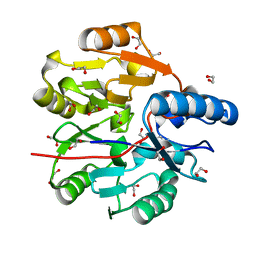 | | Crystal Structure of N-terminal domain of ArgZ, bound to Product, an arginine dihydrolase from the Ornithine-Ammonia Cycle in Cyanobacteria | | Descriptor: | 1,2-ETHANEDIOL, L-ornithine, Sll1336 protein | | Authors: | Zhuang, N, Li, L, Wu, X, Zhang, Y. | | Deposit date: | 2019-04-15 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Crystal structures and biochemical analyses of the bacterial arginine dihydrolase ArgZ suggests a "bond rotation" catalytic mechanism.
J.Biol.Chem., 295, 2020
|
|
5SHL
 
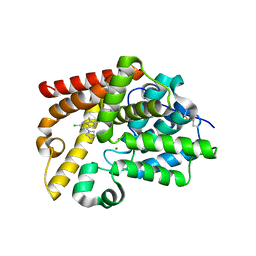 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(cc(nn2c1nc(c2C)C)Cl)N3CCCC3, micromolar IC50=1.188091 | | Descriptor: | (4R)-6-chloro-2,3-dimethyl-8-(pyrrolidin-1-yl)imidazo[1,2-b]pyridazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
2PX8
 
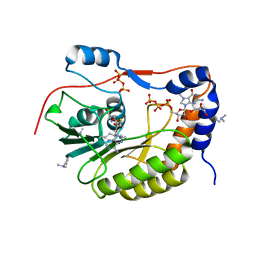 | | Crystal structure of the Murray Valley Encephalitis Virus NS5 2'-O Methyltransferase domain in complex with SAH and 7M-GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Assenberg, R, Ren, J, Verma, A, Walter, T.S, Alderton, D, Hurrelbrink, R.J, Fuller, S.D, Owens, R.J, Stuart, D.I, Grimes, J.M, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-05-14 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues.
J.Gen.Virol., 88, 2007
|
|
3KWE
 
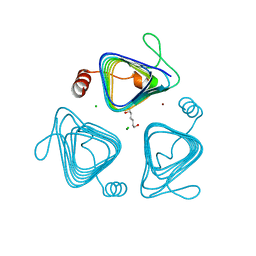 | | Inactive truncation of the beta-carboxysomal gamma-Carbonic Anhydrase, CcmM, form 2 | | Descriptor: | CHLORIDE ION, Carbon dioxide concentrating mechanism protein, HEXANE-1,6-DIOL, ... | | Authors: | Kimber, M.S, Castel, S.E, Pena, K.L. | | Deposit date: | 2009-12-01 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis of the oxidative activation of the carboxysomal {gamma}-carbonic anhydrase, CcmM.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5L62
 
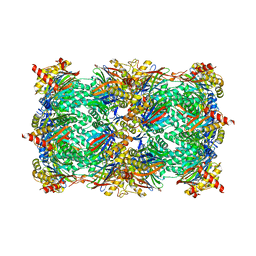 | | Yeast 20S proteasome with human beta5c (1-138) and human beta6 (97-111; 118-133) in complex with epoxyketone inhibitor 16 | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-~{N}-[(2~{S},3~{S},4~{R})-4-methyl-3,5-bis(oxidanyl)-1-phenyl-pentan-2-yl]-2-[[(2~{R})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
7AIX
 
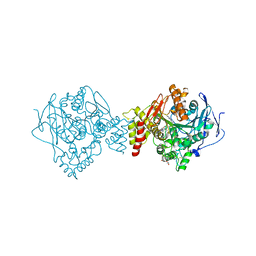 | | Crystal structure of Torpedo Californica acetylcholinesterase in complex with 2-{1-[4-(12-Amino-3-chloro-6,7,10,11-tetrahydro-7,11-methanocycloocta[b]quinolin-9-yl)butyl]-1H-1,2,3-triazol-4-yl}-N-[4-hydroxy-3-methoxybenzyl]acetamide | | Descriptor: | 2-{1-[4-(12-Amino-3-chloro-6,7,10,11-tetrahydro-7,11-methanocycloocta[b]quinolin-9-yl)butyl]-1H-1,2,3-triazol-4-yl}-N-[4-hydroxy-3-methoxybenzyl]acetamide, Acetylcholinesterase, CHLORIDE ION, ... | | Authors: | Coquelle, N, Colletier, J.P. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of a Potent Dual Inhibitor of Acetylcholinesterase and Butyrylcholinesterase with Antioxidant Activity that Alleviates Alzheimer-like Pathology in Old APP/PS1 Mice.
J.Med.Chem., 64, 2021
|
|
2PU1
 
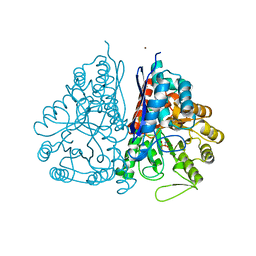 | | Crystal Structure of the T. brucei enolase complexed with Fluoro-phosphonoacetohydroxamate (FPAH) | | Descriptor: | 1,2-ETHANEDIOL, Enolase, ZINC ION, ... | | Authors: | Navarro, M.V.A.S, Rigden, D.J, Garratt, R.C, Dias, S.M.G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural flexibility in Trypanosoma brucei enolase revealed by X-ray crystallography and molecular dynamics.
Febs J., 274, 2007
|
|
5L5E
 
 | |
5L5Y
 
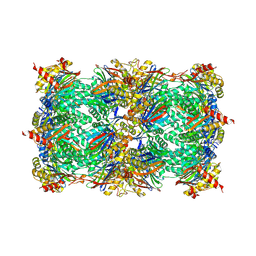 | |
6PRG
 
 | | SBP RafE in complex with stachyose | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter sugar-binding protein, CITRIC ACID, ... | | Authors: | Meier, E.P.W, Boraston, A.B. | | Deposit date: | 2019-07-10 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular analysis of an enigmaticStreptococcus pneumoniaevirulence factor: The raffinose-family oligosaccharide utilization system.
J.Biol.Chem., 294, 2019
|
|
4DMR
 
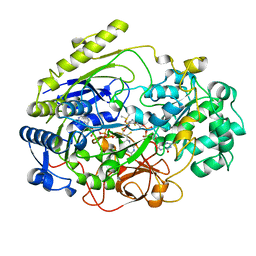 | | REDUCED DMSO REDUCTASE FROM RHODOBACTER CAPSULATUS WITH BOUND DMSO SUBSTRATE | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DIMETHYL SULFOXIDE, DMSO REDUCTASE, ... | | Authors: | Mcalpine, A.S, Bailey, S. | | Deposit date: | 1997-04-30 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The high resolution crystal structure of DMSO reductase in complex with DMSO.
J.Mol.Biol., 275, 1998
|
|
