1X8G
 
 | |
1X8H
 
 | |
1X8I
 
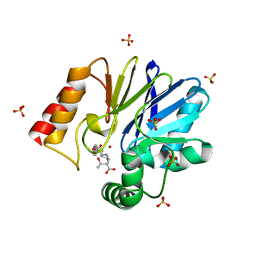 | | Crystal Structure of the Zinc Carbapenemase CphA in Complex with the Antibiotic Biapenem | | Descriptor: | 5H-PYRAZOLO(1,2-A)(1,2,4)TRIAZOL-4-IUM, 6-((2-CARBOXY-6-(1-HYDROXYETHYL)-4-METHYL-7-OXO-1-AZABICYCLO(3.2.0)HEPT-2-EN-3-YL)THIO)-6,7-DIHYDRO-, HYDROXIDE, ... | | Authors: | Garau, G, Dideberg, O. | | Deposit date: | 2004-08-18 | | Release date: | 2004-12-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Metallo-beta-lactamase Enzyme in Action: Crystal Structures of the Monozinc Carbapenemase CphA and its Complex with Biapenem
J.Mol.Biol., 345, 2005
|
|
1X8J
 
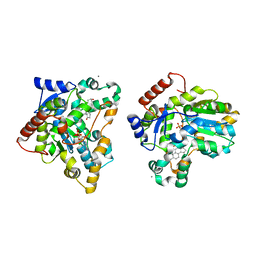 | | Crystal structure of retinol dehydratase in complex with androsterone and inactive cofactor PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Androsterone, CALCIUM ION, ... | | Authors: | Pakhomova, S, Buck, J, Newcomer, M.E. | | Deposit date: | 2004-08-18 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structures of the unique sulfotransferase retinol dehydratase with product and inhibitors provide insight into enzyme mechanism and inhibition.
Protein Sci., 14, 2005
|
|
1X8K
 
 | | Crystal structure of retinol dehydratase in complex with anhydroretinol and inactive cofactor PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, ANHYDRORETINOL, CALCIUM ION, ... | | Authors: | Pakhomova, S, Buck, J, Newcomer, M.E. | | Deposit date: | 2004-08-18 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structures of the unique sulfotransferase retinol dehydratase with product and inhibitors provide insight into enzyme mechanism and inhibition.
Protein Sci., 14, 2005
|
|
1X8L
 
 | | Crystal structure of retinol dehydratase in complex with all-trans-4-oxoretinol and inactive cofactor PAP | | Descriptor: | 4-OXORETINOL, ADENOSINE-3'-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Pakhomova, S, Buck, J, Newcomer, M.E. | | Deposit date: | 2004-08-18 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structures of the unique sulfotransferase retinol dehydratase with product and inhibitors provide insight into enzyme mechanism and inhibition.
Protein Sci., 14, 2005
|
|
1X8M
 
 | |
1X8N
 
 | | 1.08 A Crystal Structure Of Nitrophorin 4 From Rhodnius Prolixus Complexed With Nitric Oxide at pH 7.4 | | Descriptor: | NITRIC OXIDE, Nitrophorin 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kondrashov, D.A, Roberts, S.A, Weichsel, A, Montfort, W.R. | | Deposit date: | 2004-08-18 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Protein functional cycle viewed at atomic resolution: conformational change and mobility in nitrophorin 4 as a function of pH and NO binding
Biochemistry, 43, 2004
|
|
1X8O
 
 | | 1.01 A Crystal Structure Of Nitrophorin 4 From Rhodnius Prolixus Complexed With Nitric Oxide at pH 5.6 | | Descriptor: | NITRIC OXIDE, Nitrophorin 4, PHOSPHATE ION, ... | | Authors: | Kondrashov, D.A, Roberts, S.A, Weichsel, A, Montfort, W.R. | | Deposit date: | 2004-08-18 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Protein functional cycle viewed at atomic resolution: conformational change and mobility in nitrophorin 4 as a function of pH and NO binding
Biochemistry, 43, 2004
|
|
1X8P
 
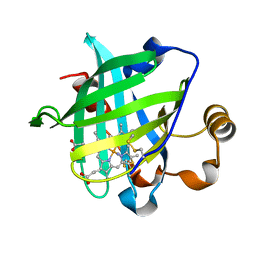 | | 0.85 A Crystal Structure Of Nitrophorin 4 From Rhodnius Prolixus Complexed With Ammonia at pH 7.4 | | Descriptor: | AMMONIA, Nitrophorin 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kondrashov, D.A, Roberts, S.A, Weichsel, A, Montfort, W.R. | | Deposit date: | 2004-08-18 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Protein functional cycle viewed at atomic resolution: conformational change and mobility in nitrophorin 4 as a function of pH and NO binding
Biochemistry, 43, 2004
|
|
1X8Q
 
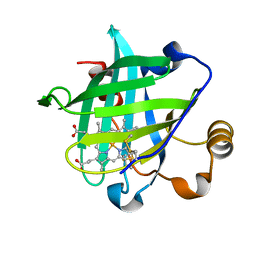 | | 0.85 A Crystal Structure Of Nitrophorin 4 From Rhodnius Prolixus in Complex with Water at pH 5.6 | | Descriptor: | Nitrophorin 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kondrashov, D.A, Roberts, S.A, Weichsel, A, Montfort, W.R. | | Deposit date: | 2004-08-18 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Protein functional cycle viewed at atomic resolution: conformational change and mobility in nitrophorin 4 as a function of pH and NO binding
Biochemistry, 43, 2004
|
|
1X8R
 
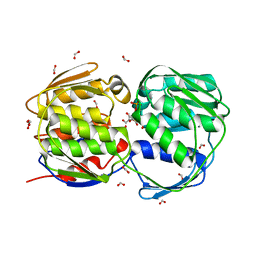 | | EPSPS liganded with the (S)-phosphonate analog of the tetrahedral reaction intermediate | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, [3R-[3A,4A,5B(S*)]]-5-(1-CARBOXY-1-PHOSPHONOETHOXY)-4-HYDROXY-3-(PHOSPHONOOXY)-1-CYCLOHEXENE-1-CARBOXYLIC ACID | | Authors: | Priestman, M.A, Healy, M.L, Becker, A, Alberg, D.G, Bartlett, P.A, Schonbrunn, E. | | Deposit date: | 2004-08-18 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interaction of phosphonate analogues of the tetrahedral reaction intermediate with 5-enolpyruvylshikimate-3-phosphate synthase in atomic detail.
Biochemistry, 44, 2005
|
|
1X8S
 
 | |
1X8T
 
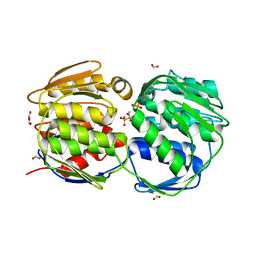 | | EPSPS liganded with the (R)-phosphonate analog of the tetrahedral reaction intermediate | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, [3R-[3A,4A,5B(R*)]]-5-(1-CARBOXY-1-PHOSPHONOETHOXY)-4-HYDROXY-3-(PHOSPHONOOXY)-1-CYCLOHEXENE-1-CARBOXYLIC ACID | | Authors: | Priestman, M.A, Healy, M.L, Becker, A, Alberg, D.G, Bartlett, P.A, Schonbrunn, E. | | Deposit date: | 2004-08-18 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interaction of phosphonate analogues of the tetrahedral reaction intermediate with 5-enolpyruvylshikimate-3-phosphate synthase in atomic detail.
Biochemistry, 44, 2005
|
|
1X8U
 
 | | Crystal structure of Siderocalin (NGAL, Lipocalin 2) complexed with Carboxymycobactin T | | Descriptor: | CARBOXYMYCOBACTIN T, Neutrophil gelatinase-associated lipocalin | | Authors: | Holmes, M.A, Paulsene, W, Jide, X, Ratledge, C, Strong, R.K. | | Deposit date: | 2004-08-18 | | Release date: | 2005-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Siderocalin (Lcn 2) Also Binds Carboxymycobactins, Potentially Defending against Mycobacterial Infections through Iron Sequestration
Structure, 13, 2005
|
|
1X8V
 
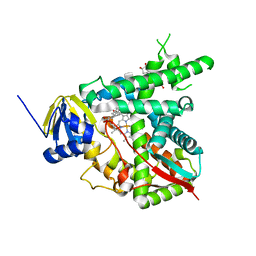 | | Estriol-bound and ligand-free structures of sterol 14alpha-demethylase (CYP51) | | Descriptor: | Cytochrome P450 51, ESTRIOL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Podust, L.M, Yermalitskaya, L.V, Lepesheva, G.I, Podust, V.N, Dalmasso, E.A, Waterman, M.R. | | Deposit date: | 2004-08-18 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Estriol Bound and Ligand-free Structures of Sterol 14alpha-Demethylase.
Structure, 12, 2004
|
|
1X8W
 
 | |
1X8X
 
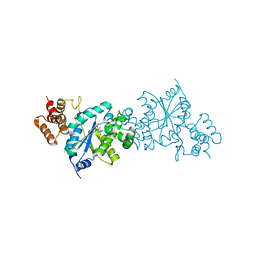 | | Tyrosyl t-RNA Synthetase from E.coli Complexed with Tyrosine | | Descriptor: | SULFATE ION, TYROSINE, Tyrosyl-tRNA synthetase | | Authors: | Kobayashi, T, Takimura, T, Sekine, R, Kelly, V.P, Kamata, K, Sakamoto, K, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-08-19 | | Release date: | 2005-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Snapshots of the KMSKS Loop Rearrangement for Amino Acid Activation by Bacterial Tyrosyl-tRNA Synthetase
J.MOL.BIOL., 346, 2005
|
|
1X8Y
 
 | | Human lamin coil 2B | | Descriptor: | Lamin A/C | | Authors: | Strelkov, S.V, Schumacher, J, Burkhard, P, Aebi, U, Herrmann, H. | | Deposit date: | 2004-08-19 | | Release date: | 2004-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the human lamin A coil 2B dimer: implications for the head-to-tail association of nuclear lamins
J.Mol.Biol., 343, 2004
|
|
1X8Z
 
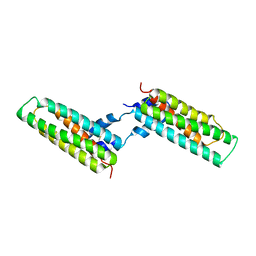 | | Crystal structure of a pectin methylesterase inhibitor from Arabidopsis thaliana | | Descriptor: | invertase/pectin methylesterase inhibitor family protein | | Authors: | Hothorn, M, Wolf, S, Aloy, P, Greiner, S, Scheffzek, K. | | Deposit date: | 2004-08-19 | | Release date: | 2004-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural insights into the target specificity of plant invertase and pectin methylesterase inhibitory proteins
Plant Cell, 16, 2004
|
|
1X90
 
 | | Crystal structure of mutant form B of a pectin methylesterase inhibitor from Arabidopsis | | Descriptor: | invertase/pectin methylesterase inhibitor family protein | | Authors: | Hothorn, M, Wolf, S, Aloy, P, Greiner, S, Scheffzek, K. | | Deposit date: | 2004-08-19 | | Release date: | 2004-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural insights into the target specificity of plant invertase and pectin methylesterase inhibitory proteins
Plant Cell, 16, 2004
|
|
1X91
 
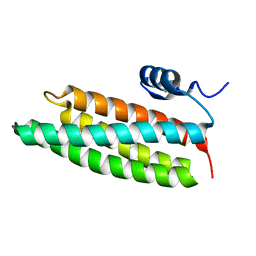 | | Crystal structure of mutant form A of a pectin methylesterase inhibitor from Arabidopsis | | Descriptor: | invertase/pectin methylesterase inhibitor family protein | | Authors: | Hothorn, M, Wolf, S, Aloy, P, Greiner, S, Scheffzek, K. | | Deposit date: | 2004-08-19 | | Release date: | 2004-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the target specificity of plant invertase and pectin methylesterase inhibitory proteins
Plant Cell, 16, 2004
|
|
1X92
 
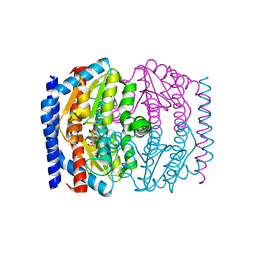 | | CRYSTAL STRUCTURE OF PSEUDOMONAS AERUGINOSA PHOSPHOHEPTOSE ISOMERASE IN COMPLEX WITH REACTION PRODUCT D-GLYCERO-D-MANNOPYRANOSE-7-PHOSPHATE | | Descriptor: | 7-O-phosphono-D-glycero-alpha-D-manno-heptopyranose, PHOSPHOHEPTOSE ISOMERASE | | Authors: | Walker, J.R, Evdokimova, E, Kudritska, M, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-19 | | Release date: | 2004-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of sedoheptulose-7-phosphate isomerase, a critical enzyme for lipopolysaccharide biosynthesis and a target for antibiotic adjuvants.
J.Biol.Chem., 283, 2008
|
|
1X93
 
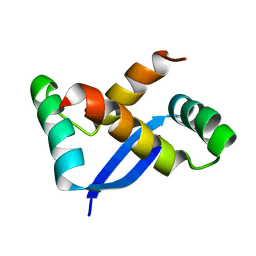 | | NMR Structure of Helicobacter pylori HP0222 | | Descriptor: | hypothetical protein HP0222 | | Authors: | Popescu, A, Karpay, A, Israel, D, Peek Jr, R.M, Krezel, A.M. | | Deposit date: | 2004-08-19 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Helicobacter pylori protein HP0222 belongs to Arc/MetJ family of transcriptional regulators.
Proteins, 59, 2005
|
|
1X94
 
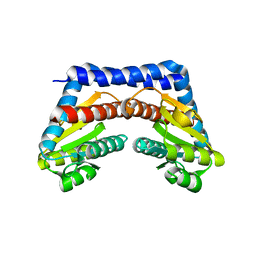 | |
