7L3D
 
 | | T4 Lysozyme L99A - 3-iodotoluene - RT | | Descriptor: | 1-iodo-3-methylbenzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
5CAS
 
 | | EGFR kinase domain mutant "TMLR" with compound 41a | | Descriptor: | (1R)-1-{6-({2-[(3R,4S)-3-fluoro-4-methoxypiperidin-1-yl]pyrimidin-4-yl}amino)-1-[(2S)-1,1,1-trifluoropropan-2-yl]-1H-imidazo[4,5-c]pyridin-2-yl}ethanol, Epidermal growth factor receptor, SULFATE ION | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2015-06-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Noncovalent Mutant Selective Epidermal Growth Factor Receptor Inhibitors: A Lead Optimization Case Study.
J.Med.Chem., 58, 2015
|
|
7L3E
 
 | | T4 Lysozyme L99A - 3-iodotoluene - cryo | | Descriptor: | 1-iodo-3-methylbenzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7QYR
 
 | | Crystal structure of RimK from Pseudomonas aeruginosa PAO1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Probable alpha-L-glutamate ligase, poly-glutamate | | Authors: | Thompson, C.M.A, Little, R.H, Stevenson, C.E.M, Lawson, D.M, Malone, J.G. | | Deposit date: | 2022-01-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the mechanism of adaptive ribosomal modification by Pseudomonas RimK.
Proteins, 91, 2023
|
|
4NB7
 
 | | Crystal Structure of Two-Domain Laccase from Streptomyces LIvidans AC1709 in complex with azide after 180 min soaking | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, AZIDE ION, ... | | Authors: | Gabdulkhakov, A, Tischenko, S, Yurevich, L, Lisov, A, Leontievsky, A. | | Deposit date: | 2013-10-23 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of Two-Domain Laccase from Streptomyces Lividans AC1709 in complex with azide after 180 min soaking
To be Published
|
|
7L3I
 
 | | T4 Lysozyme L99A - propylbenzene - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Endolysin, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
4MWQ
 
 | | Anhui N9-oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
4NBF
 
 | | Oxygenase with Gln282 replaced by Asn and ferredoxin complex of carbazole 1,9a-dioxygenase | | Descriptor: | FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin CarAc, ... | | Authors: | Ashikawa, Y, Usami, Y, Inoue, K, Nojiri, H. | | Deposit date: | 2013-10-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the divergent oxygenation reactions catalyzed by the rieske nonheme iron oxygenase carbazole 1,9a-dioxygenase.
Appl.Environ.Microbiol., 80, 2014
|
|
2E3T
 
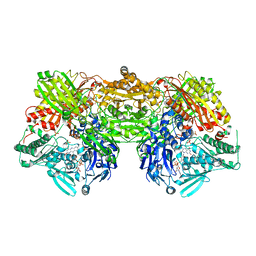 | | Crystal structure of rat xanthine oxidoreductase mutant (W335A and F336L) | | Descriptor: | BICARBONATE ION, CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Asai, R, Nishino, T, Matsumura, T, Okamoto, K, Pai, E.F, Nishino, T. | | Deposit date: | 2006-11-28 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Two mutations convert mammalian xanthine oxidoreductase to highly superoxide-productive xanthine oxidase
J.Biochem.(Tokyo), 141, 2007
|
|
2YDF
 
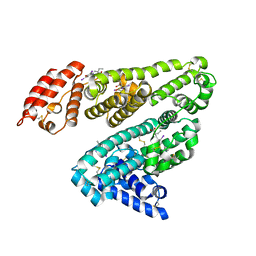 | | HUMAN SERUM ALBUMIN COMPLEXED WITH IOPHENOXIC ACID | | Descriptor: | IOPHENOXIC ACID, SERUM ALBUMIN | | Authors: | Ryan, A.J, Curry, S. | | Deposit date: | 2011-03-18 | | Release date: | 2011-04-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystallographic Analysis Reveals the Structural Basis of the High-Affinity Binding of Iophenoxic Acid to Human Serum Albumin.
Bmc Struct.Biol., 11, 2011
|
|
4MXJ
 
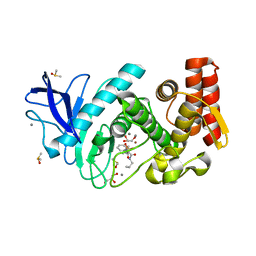 | | Thermolysin in complex with UBTLN35 | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2013-09-26 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Methyl, Ethyl, Propyl, Butyl: Futile But Not for Water, as the Correlation of Structure and Thermodynamic Signature Shows in a Congeneric Series of Thermolysin Inhibitors.
Chemmedchem, 4, 2014
|
|
3DAP
 
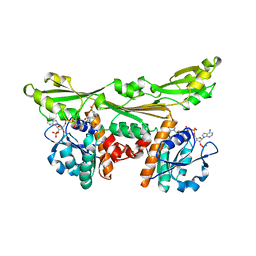 | | C. GLUTAMICUM DAP DEHYDROGENASE IN COMPLEX WITH NADP+ AND THE INHIBITOR 5S-ISOXAZOLINE | | Descriptor: | (2S,5',S)-2-AMINO-3-(3-CARBOXY-2-ISOXAZOLIN-5-YL)PROPANOIC ACID, DIAMINOPIMELIC ACID DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Scapin, G, Cirilli, M, Reddy, S.G, Gao, Y, Vederas, J.C, Blanchard, J.S. | | Deposit date: | 1997-12-29 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate and inhibitor binding sites in Corynebacterium glutamicum diaminopimelate dehydrogenase.
Biochemistry, 37, 1998
|
|
2YQ0
 
 | |
2YBU
 
 | | Crystal structure of human acidic chitinase in complex with bisdionin F | | Descriptor: | 3,7-DIMETHYL-1-[3-(3-METHYL-2,6-DIOXO-9H-PURIN-1-YL)PROPYL]PURINE-2,6-DIONE, ACIDIC MAMMALIAN CHITINASE, GLYCEROL | | Authors: | Sutherland, T.E, Andersen, O.A, Betou, M, Eggleston, I.M, Maizels, R.M, van Aalten, D, Allen, J.E. | | Deposit date: | 2011-03-10 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Analyzing airway inflammation with chemical biology: dissection of acidic mammalian chitinase function with a selective drug-like inhibitor.
Chem. Biol., 18, 2011
|
|
8UQW
 
 | | Round 18 Arylesterase Variant of Apo-Phosphotriesterase Measured at 13 keV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphotriesterase variant PTE-R18 | | Authors: | Breeze, C.W, Frkic, R.L, Campbell, E.C, Jackson, C.J. | | Deposit date: | 2023-10-25 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mononuclear binding and catalytic activity of europium(III) and gadolinium(III) at the active site of the model metalloenzyme phosphotriesterase.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
4H3E
 
 | |
1PH0
 
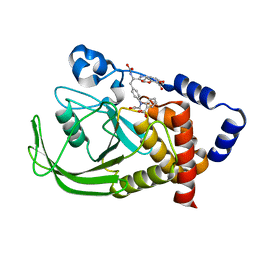 | | Non-carboxylic Acid-Containing Inhibitor of PTP1B Targeting the Second Phosphotyrosine Site | | Descriptor: | 2-{4-[2-(S)-ALLYLOXYCARBONYLAMINO-3-{4-[(2-CARBOXY-PHENYL)-OXALYL-AMINO]-PHENYL}-PROPIONYLAMINO]-BUTOXY}-6-HYDROXY-BENZ OIC ACID METHYL ESTER, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Liu, G, Xin, Z, Liang, H, Abad-Zapatero, C, Hajduk, P, Janowick, D, Szczepankiewicz, B, Pei, Z, Hutchins, C.W, Ballaron, S.J. | | Deposit date: | 2003-05-29 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selective Protein Tyrosine Phosphatase 1B Inhibitors: Targeting the Second Phosphotyrosine Binding Site with Non-Carboxylic Acid-Containing Ligands.
J.Med.Chem., 46, 2003
|
|
7QHL
 
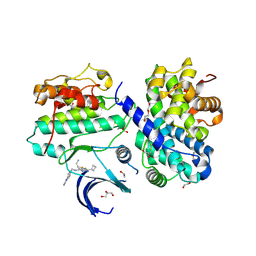 | | Crystal structure of Cyclin-dependent kinase 2/cyclin A in complex with 3,5,7-Substituted pyrazolo[4,3-d]pyrimidine inhibitor 24 | | Descriptor: | 1,2-ETHANEDIOL, 5-(2-amino-1-ethyl)thio-3-cyclobutyl-7-[4-(pyrazol-1-yl)benzyl]amino-1(2)H-pyrazolo[4,3-d]pyrimidine, Cyclin-A2, ... | | Authors: | Djukic, S, Skerlova, J, Rezacova, P. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 3,5,7-Substituted Pyrazolo[4,3- d ]Pyrimidine Inhibitors of Cyclin-Dependent Kinases and Cyclin K Degraders.
J.Med.Chem., 65, 2022
|
|
7QYS
 
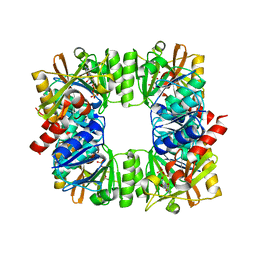 | | Crystal structure of RimK from Pseudomonas syringae DC3000 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Probable alpha-L-glutamate ligase | | Authors: | Thompson, C.M.A, Little, R.H, Stevenson, C.E.M, Lawson, D.M, Malone, J.G. | | Deposit date: | 2022-01-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the mechanism of adaptive ribosomal modification by Pseudomonas RimK.
Proteins, 91, 2023
|
|
4MY7
 
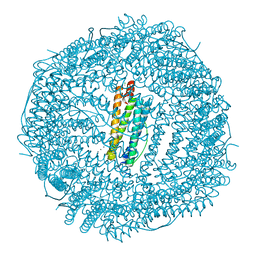 | | frog M ferritin iron-loaded under anaerobic environment | | Descriptor: | CHLORIDE ION, FE (II) ION, Ferritin, ... | | Authors: | Mangani, S, Di Pisa, F, Pozzi, C, Turano, P, Lalli, D. | | Deposit date: | 2013-09-27 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Time-lapse anomalous X-ray diffraction shows how Fe(2+) substrate ions move through ferritin protein nanocages to oxidoreductase sites.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1PHB
 
 | | INHIBITOR-INDUCED CONFORMATIONAL CHANGE IN CYTOCHROME P450-CAM | | Descriptor: | 1-(N-IMIDAZOLYL)-2-HYDROXY-2-(2,3-DICHLOROPHENYL)OCTANE, CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Poulos, T.L. | | Deposit date: | 1992-07-27 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibitor-induced conformational change in cytochrome P-450CAM.
Biochemistry, 32, 1993
|
|
2YEY
 
 | | Crystal structure of the allosteric-defective chaperonin GroEL E434K mutant | | Descriptor: | 60 KDA CHAPERONIN | | Authors: | Cabo-Bilbao, A, Mechaly, A.E, Agirre, J, Spinelli, S, Sot, B, Muga, A, Guerin, D.M.A. | | Deposit date: | 2011-03-31 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Crystal Structure of the Temperature-Sensitive and Allosteric-Defective Chaperonin Groele461K.
J.Struct.Biol., 155, 2006
|
|
5FZ0
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with 2,5-dichloro-N-(pyridin-3-yl)thiophene-3-carboxamide (N08137b) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 2,5-dichloro-N-(pyridin-3-yl)thiophene-3-carboxamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Nowak, R, Krojer, T, Pearce, N, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-10 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with N11213A
To be Published
|
|
3DA7
 
 | | A conformationally strained, circular permutant of barnase | | Descriptor: | Barnase circular permutant, Barstar | | Authors: | Mitrousis, G, Butler, J, Loh, S.N, Cingolani, G. | | Deposit date: | 2008-05-28 | | Release date: | 2009-04-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and thermodynamic analysis of a conformationally strained circular permutant of barnase.
Biochemistry, 48, 2009
|
|
8UQY
 
 | | Round 18 Arylesterase Variant of Phosphotriesterase Bound to Europium(III) Measured at 9.5 keV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, EUROPIUM (III) ION, ... | | Authors: | Breeze, C.W, Frkic, R.L, Campbell, E.C, Jackson, C.J. | | Deposit date: | 2023-10-25 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mononuclear binding and catalytic activity of europium(III) and gadolinium(III) at the active site of the model metalloenzyme phosphotriesterase.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
