4YVI
 
 | |
9I0T
 
 | | Crystal structure of TRIM25 PRYSPRY covalently bound to 2-chloro-1-[4-(3-methyl-4-phenyl-phenyl)carbonyl-1,4-diazepan-1-yl]ethanone | | Descriptor: | 1-[4-(3-methyl-4-phenyl-phenyl)carbonyl-1,4-diazepan-1-yl]ethanone, E3 ubiquitin/ISG15 ligase TRIM25 | | Authors: | McPhie, K.A, Esposito, D, Rittinger, K. | | Deposit date: | 2025-01-15 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and optimisation of a covalent ligand for TRIM25 and its application to targeted protein ubiquitination.
Chem Sci, 16, 2025
|
|
5EFN
 
 | |
4NDW
 
 | |
5V0E
 
 | | Crystal structure of human exonuclease 1 Exo1 (WT) in complex with 5' flap DNA (f5I) | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*AP*GP*TP*GP*AP*TP*AP*C)-3'), DNA (5'-D(P*CP*TP*CP*GP*TP*CP*AP*CP*TP*AP*GP*CP*G)-3'), Exonuclease 1, ... | | Authors: | Shi, Y, Beese, L.S. | | Deposit date: | 2017-02-28 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.744 Å) | | Cite: | Interplay of catalysis, fidelity, threading, and processivity in the exo- and endonucleolytic reactions of human exonuclease I.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5EH5
 
 | | human carbonic anhydrase II in complex with ligand | | Descriptor: | 1.7.6 4-methylpyrimidine-2-sulfonamide, Carbonic anhydrase 2, FORMIC ACID, ... | | Authors: | Ren, B. | | Deposit date: | 2015-10-28 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.207 Å) | | Cite: | Native State Mass Spectrometry, Surface Plasmon Resonance, and X-ray Crystallography Correlate Strongly as a Fragment Screening Combination.
J.Med.Chem., 59, 2016
|
|
4I17
 
 | |
5EHR
 
 | |
6JAP
 
 | | Crystal structure of ABC transporter alpha-glycoside-binding mutant protein R356A in complex with sucrose | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, periplasmic substrate-binding protein, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
5AYD
 
 | | Crystal structure of Ruminococcus albus beta-(1,4)-mannooligosaccharide phosphorylase (RaMP2) in complexes with phosphate | | Descriptor: | Beta-1,4-mannooligosaccharide phosphorylase, PHOSPHATE ION | | Authors: | Ye, Y, Saburi, W, Kato, K, Yao, M. | | Deposit date: | 2015-08-13 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the difference in substrate recognition of two mannoside phosphorylases from two GH130 subfamilies.
Febs Lett., 590, 2016
|
|
5EHN
 
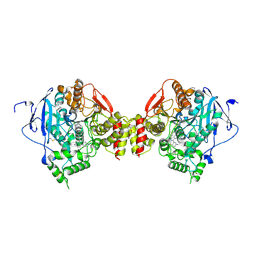 | | mAChE-syn TZ2PA5 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-phenyl-5-[5-[3-[2-(1,2,3,4-tetrahydroacridin-9-ylamino)ethyl]-1,2,3-triazol-4-yl]pentyl]phenanthridin-5-ium-3,8-diamine, Acetylcholinesterase, ... | | Authors: | Bourne, Y, Marchot, P. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Steric and Dynamic Parameters Influencing In Situ Cycloadditions to Form Triazole Inhibitors with Crystalline Acetylcholinesterase.
J.Am.Chem.Soc., 138, 2016
|
|
6J20
 
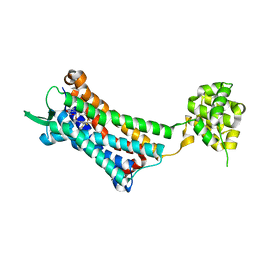 | | Crystal structure of the human NK1 substance P receptor | | Descriptor: | 5-[[(2~{R},3~{S})-2-[(1~{R})-1-[3,5-bis(trifluoromethyl)phenyl]ethoxy]-3-(4-fluorophenyl)morpholin-4-yl]methyl]-1,2-dihydro-1,2,4-triazol-3-one, Substance-P receptor,Endolysin | | Authors: | Chen, S, Lu, M, Zhang, H, Wu, B, Zhao, Q. | | Deposit date: | 2018-12-30 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human substance P receptor binding mode of the antagonist drug aprepitant by NMR and crystallography.
Nat Commun, 10, 2019
|
|
5EJ0
 
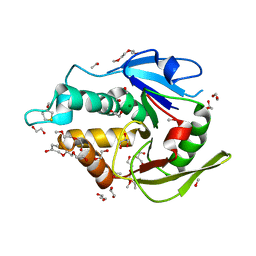 | | The vaccinia virus H3 envelope protein, a major target of neutralizing antibodies, exhibits a glycosyltransferase fold and binds UDP-Glucose | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Singh, K, Gittis, A.G, Gitti, R.K, Ostazesky, S.A, Su, H.P, Garboczi, D.N. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Vaccinia Virus H3 Envelope Protein, a Major Target of Neutralizing Antibodies, Exhibits a Glycosyltransferase Fold and Binds UDP-Glucose.
J.Virol., 90, 2016
|
|
5HVK
 
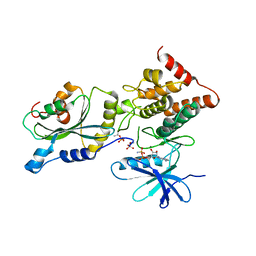 | |
2VAG
 
 | | Crystal structure of di-phosphorylated human CLK1 in complex with a novel substituted indole inhibitor | | Descriptor: | DUAL SPECIFICITY PROTEIN KINASE CLK1, ethyl 3-[(E)-2-amino-1-cyanoethenyl]-6,7-dichloro-1-methyl-1H-indole-2-carboxylate | | Authors: | Pike, A.C.W, Bullock, A.N, Fedorov, O, Pilka, E.S, Ugochukwu, E, von Delft, F, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Huber, K, Bracher, F, Knapp, S. | | Deposit date: | 2007-08-31 | | Release date: | 2007-10-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Specific Clk Inhibitors from a Novel Chemotype for Regulation of Alternative Splicing.
Chem.Biol, 18, 2011
|
|
5ZLM
 
 | | Mutation in the trinuclear site of CotA-laccase: H491C mutant, PH 8.0 | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Xie, T, Liu, Z.C, Wang, G.G. | | Deposit date: | 2018-03-28 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insight into the Allosteric Coupling of Cu1 Site and Trinuclear Cu Cluster in CotA Laccase.
Chembiochem, 19, 2018
|
|
4AE3
 
 | | Crystal structure of ammosamide 272:myosin-2 motor domain complex | | Descriptor: | 1,2-ETHANEDIOL, ADP ORTHOVANADATE, AMMOSAMIDE 272, ... | | Authors: | Chinthalapudi, K, Heissler, S.M, Fenical, W, Manstein, D.J. | | Deposit date: | 2012-01-05 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Ammosamide Mediated Myosin Motor Activity Inhibition
To be Published
|
|
3MCJ
 
 | | Crystal structure of molybdenum cofactor biosynthesis (AQ_061) other form from aquifex aeolicus VF5 | | Descriptor: | 1,2-ETHANEDIOL, Molybdenum cofactor biosynthesis MOG | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-03-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures, dynamics and functional implications of molybdenum-cofactor biosynthesis protein MogA from two thermophilic organisms
Acta Crystallogr.,Sect.F, 67, 2011
|
|
5C0V
 
 | | Structure of the LARP1-unique domain DM15 | | Descriptor: | La-related protein 1, SULFATE ION | | Authors: | Lahr, R.M, Berman, A.J. | | Deposit date: | 2015-06-12 | | Release date: | 2015-08-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The La-related protein 1-specific domain repurposes HEAT-like repeats to directly bind a 5'TOP sequence.
Nucleic Acids Res., 43, 2015
|
|
2VD8
 
 | | The crystal structure of alanine racemase from Bacillus anthracis (BA0252) | | Descriptor: | ALANINE RACEMASE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Au, K, Ren, J, Walter, T.S, Harlos, K, Nettleship, J.E, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-10-01 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structures of an Alanine Racemase from Bacillus Anthracis (Ba0252) in the Presence and Absence of (R)-1-Aminoethylphosphonic Acid (L-Ala-P).
Acta Crystallogr.,Sect.F, 64, 2008
|
|
5ZLK
 
 | | Mutation in the trinuclear site of CotA-laccase: H493A mutant, PH 8.0 | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Xie, T, Liu, Z.C, Wang, G.G. | | Deposit date: | 2018-03-28 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insight into the Allosteric Coupling of Cu1 Site and Trinuclear Cu Cluster in CotA Laccase.
Chembiochem, 19, 2018
|
|
1D79
 
 | | HIGH RESOLUTION REFINEMENT OF THE HEXAGONAL A-DNA OCTAMER D(GTGTACAC) AT 1.4 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*AP*CP*AP*C)-3') | | Authors: | Thota, N, Li, X.H, Bingman, C.A, Sundaralingam, M. | | Deposit date: | 1992-06-12 | | Release date: | 1993-04-15 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution refinement of the hexagonal A-DNA octamer d(GTGTACAC) at 1.4 A.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
4K2Z
 
 | | Crystal structure of the complex of type I Ribosome inactivating protein from Momordica balsamina with Methylethylamine at 1.80 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, METHYLETHYLAMINE, ... | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the complex of type I Ribosome inactivating protein from Momordica balsamina with Methylethylamine at 1.80 A resolution
To be Published
|
|
7CRU
 
 | | hnRNPK NLS in complex with Importin alpha 1 (KPNA2) | | Descriptor: | ACETATE ION, GLYCEROL, Heterogeneous nuclear ribonucleoprotein K, ... | | Authors: | Yao, J, Sun, Q. | | Deposit date: | 2020-08-14 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nuclear import receptors and hnRNPK mediates nuclear import and stress granule localization of SIRLOIN.
Cell.Mol.Life Sci., 78, 2021
|
|
6GBX
 
 | | Crystal structure of human glutaminyl cyclase variant Y115E-Y117E in complex with SEN177 | | Descriptor: | 1,2-ETHANEDIOL, 2-fluoranyl-5-[2-[4-(4-methyl-1,2,4-triazol-3-yl)piperidin-1-yl]pyridin-3-yl]pyridine, Glutaminyl-peptide cyclotransferase, ... | | Authors: | Pozzi, C, Di Pisa, F, Benvenuti, M, Mangani, S. | | Deposit date: | 2018-04-16 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The structure of the human glutaminyl cyclase-SEN177 complex indicates routes for developing new potent inhibitors as possible agents for the treatment of neurological disorders.
J. Biol. Inorg. Chem., 23, 2018
|
|
