1L90
 
 | |
1L95
 
 | |
4O46
 
 | | 14-3-3-gamma in complex with influenza NS1 C-terminal tail phosphorylated at S228 | | Descriptor: | 14-3-3 protein gamma, Nonstructural protein 1, UNKNOWN ATOM OR ION, ... | | Authors: | Qin, S, Liu, Y, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for histone mimicry and hijacking of host proteins by influenza virus protein NS1.
Nat Commun, 5, 2014
|
|
3BZK
 
 | |
1GNT
 
 | | Hybrid Cluster Protein from Desulfovibrio vulgaris. X-ray structure at 1.25A resolution using synchrotron radiation. | | Descriptor: | HYBRID CLUSTER PROTEIN, IRON/SULFUR CLUSTER, IRON/SULFUR/OXYGEN HYBRID CLUSTER | | Authors: | Macedo, S, Mitchell, E.P, Romao, C.V, Cooper, S.J, Coelho, R, Liu, M.Y, Xavier, A.V, Legall, J, Bailey, S, Garner, D.C, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hybrid cluster proteins (HCPs) from Desulfovibrio desulfuricans ATCC 27774 and Desulfovibrio vulgaris (Hildenborough): X-ray structures at 1.25 A resolution using synchrotron radiation.
J. Biol. Inorg. Chem., 7, 2002
|
|
2FEM
 
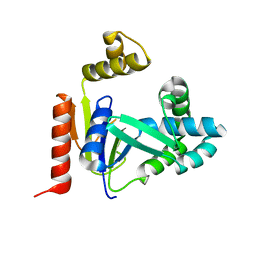 | | Mutant R188M of the Cytidine Monophosphate Kinase From E. Coli | | Descriptor: | Cytidylate kinase | | Authors: | Ofiteru, A, Bucurenci, N, Alexov, E, Bertrand, T, Briozzo, P, Munier-Lehmann, H, Tourneux, L, Barzu, O, Gilles, A.M. | | Deposit date: | 2005-12-16 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional consequences of single amino acid substitutions in the pyrimidine base binding pocket of Escherichia coli CMP kinase.
Febs J., 274, 2007
|
|
1LE7
 
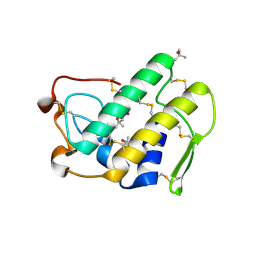 | | CARBOXYLIC ESTER HYDROLASE, C 2 2 21 space group | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Group X Secretory Phospholipase A2 | | Authors: | Pan, Y.H, Jain, M.K, Bahnson, B.J. | | Deposit date: | 2002-04-09 | | Release date: | 2002-08-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of human group X secreted phospholipase A2. Electrostatically neutral interfacial surface targets zwitterionic membranes.
J.Biol.Chem., 277, 2002
|
|
1LMC
 
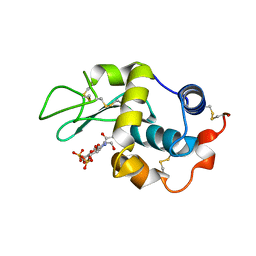 | | THE CRYSTAL STRUCTURE OF A COMPLEX BETWEEN BULGECIN, A BACTERIAL METABOLITE, AND LYSOZYME FROM THE RAINBOW TROUT | | Descriptor: | BULGECIN A, LYSOZYME | | Authors: | Karlsen, S, Hough, E. | | Deposit date: | 1994-11-14 | | Release date: | 1996-01-01 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a complex between bulgecin, a bacterial metabolite, and lysozyme from the rainbow trout.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1GX1
 
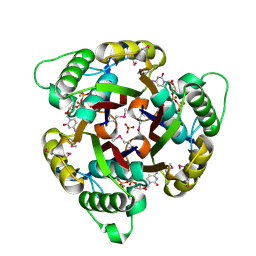 | | Structure of 2C-Methyl-D-erythritol-2,4-cyclodiphosphate Synthase | | Descriptor: | 2-C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE SYNTHASE, CYTIDINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Kemp, L.E, Bond, C.S, Hunter, W.N. | | Deposit date: | 2002-03-26 | | Release date: | 2002-04-03 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of 2C-Methyl-D-Erythritol 2,4-Cyclodiphosphate Synthase: An Essential Enzyme for Isoprenoid Biosynthesis and Target for Antimicrobial Drug Development
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LEH
 
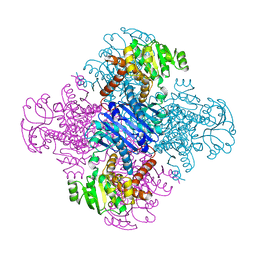 | | LEUCINE DEHYDROGENASE FROM BACILLUS SPHAERICUS | | Descriptor: | LEUCINE DEHYDROGENASE | | Authors: | Baker, P.J, Turnbull, A.P, Sedelnikova, S.E, Stillman, T.J, Rice, D.W. | | Deposit date: | 1995-06-09 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A role for quaternary structure in the substrate specificity of leucine dehydrogenase.
Structure, 3, 1995
|
|
1GUY
 
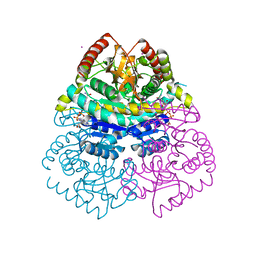 | | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases | | Descriptor: | CADMIUM ION, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dalhus, B, Sarinen, M, Sauer, U.H, Eklund, P, Johansson, K, Karlsson, A, Ramaswamy, S, Bjork, A, Synstad, B, Naterstad, K, Sirevag, R, Eklund, H. | | Deposit date: | 2002-02-04 | | Release date: | 2002-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases
J.Mol.Biol., 318, 2002
|
|
1L42
 
 | |
232D
 
 | |
1LHI
 
 | | ROLE OF PROLINE RESIDUES IN HUMAN LYSOZYME STABILITY: A SCANNING CALORIMETRIC STUDY COMBINED WITH X-RAY STRUCTURE ANALYSIS OF PROLINE MUTANTS | | Descriptor: | HUMAN LYSOZYME | | Authors: | Inaka, K, Matsushima, M, Herning, T, Kuroki, R, Yutani, K, Kikuchi, M. | | Deposit date: | 1992-03-27 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of proline residues in human lysozyme stability: a scanning calorimetric study combined with X-ray structure analysis of proline mutants.
Biochemistry, 31, 1992
|
|
1L4A
 
 | | X-RAY STRUCTURE OF THE NEURONAL COMPLEXIN/SNARE COMPLEX FROM THE SQUID LOLIGO PEALEI | | Descriptor: | S-SNAP25 fusion protein, S-SYNTAXIN, SYNAPHIN A, ... | | Authors: | Bracher, A, Kadlec, J, Betz, H, Weissenhorn, W. | | Deposit date: | 2002-03-04 | | Release date: | 2002-07-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | X-ray structure of a neuronal complexin-SNARE complex from squid.
J.Biol.Chem., 277, 2002
|
|
1H2G
 
 | | Altered substrate specificity mutant of penicillin acylase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | McVey, C.E, Morillas, M, Brannigan, J.A, Ladurner, A.G, Forney, L.J, Virden, R. | | Deposit date: | 2002-08-08 | | Release date: | 2003-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations of Penicillin Acylase Residue B71 Extend Substrate Specificity by Decreasing Steric Constraints for Substrate Binding
Biochem.J., 371, 2003
|
|
1L59
 
 | |
1H33
 
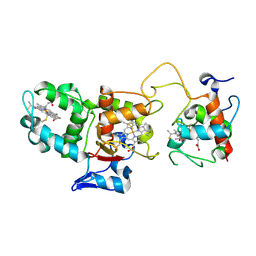 | | Oxidised SoxAX complex from Rhodovulum sulfidophilum | | Descriptor: | CYTOCHROME C, DIHEME CYTOCHROME C, HEME C | | Authors: | Bamford, V.A, Bruno, S, Rasmussen, T, Appia-Ayme, C, Cheesman, M.R, Berks, B.C, Hemmings, A.M. | | Deposit date: | 2002-08-21 | | Release date: | 2002-11-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for the Oxidation of Thiosulfate by a Sulfur Cycle Enzyme
Embo J., 21, 2002
|
|
4O55
 
 | |
1LM1
 
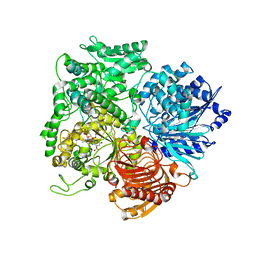 | | Structural studies on the synchronization of catalytic centers in glutamate synthase: native enzyme | | Descriptor: | ACETATE ION, FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | van Den Heuvel, R.H, Ferrari, D, Bossi, R.T, Ravasio, S, Curti, B, Vanoni, M.A, Florencio, F.J, Mattevi, A. | | Deposit date: | 2002-04-30 | | Release date: | 2002-07-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies on the synchronization of catalytic centers in glutamate synthase
J.BIOL.CHEM., 277, 2002
|
|
3AB9
 
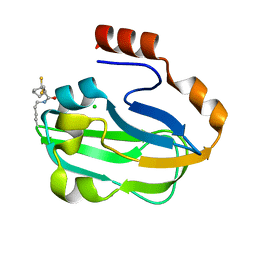 | | Crystal Structure of lipoylated E. coli H-protein (reduced form) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Glycine cleavage system H protein | | Authors: | Okamura-Ikeda, K, Maita, N. | | Deposit date: | 2009-12-04 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of aminomethyltransferase in complex with dihydrolipoyl-H-protein of the glycine cleavage system: implications for recognition of lipoyl protein substrate, disease-related mutations, and reaction mechanism
J.Biol.Chem., 285, 2010
|
|
1L70
 
 | |
1L75
 
 | |
3ADC
 
 | | Crystal structure of O-phosphoseryl-tRNA kinase complexed with selenocysteine tRNA and AMPPNP (crystal type 2) | | Descriptor: | L-seryl-tRNA(Sec) kinase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Itoh, Y, Chiba, S, Sekine, S, Yokoyama, S. | | Deposit date: | 2010-01-18 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for the Major Role of O-Phosphoseryl-tRNA Kinase in the UGA-Specific Encoding of Selenocysteine
Mol.Cell, 39, 2010
|
|
4OB4
 
 | | Structure of the S. venezulae BldD DNA-binding domain | | Descriptor: | Putative DNA-binding protein | | Authors: | schumacher, M.A, Tschowri, N, Buttner, M, Brennan, R. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development.
Cell(Cambridge,Mass.), 158, 2014
|
|
