4E2Z
 
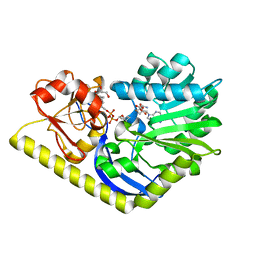 | | X-ray Structure of the H225N mutant of TcaB9, a C-3'-Methyltransferase, in Complex with S-Adenosyl-L-Homocysteine and Sugar Product | | Descriptor: | (2R,4S,6R)-4-amino-4,6-dimethyl-5-oxotetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Bruender, N.A, Holden, H.M. | | Deposit date: | 2012-03-09 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Probing the catalytic mechanism of a C-3'-methyltransferase involved in the biosynthesis of D-tetronitrose.
Protein Sci., 21, 2012
|
|
4R6X
 
 | |
2AEU
 
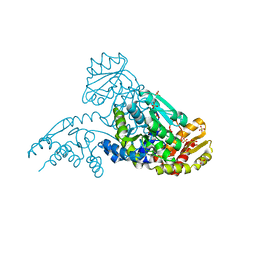 | | MJ0158, apo form | | Descriptor: | Hypothetical protein MJ0158, SULFATE ION | | Authors: | Kaiser, J.T, Gromadski, K, Rother, M, Engelhardt, H, Rodnina, M.V, Wahl, M.C. | | Deposit date: | 2005-07-24 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional investigation of a putative archaeal selenocysteine synthase
Biochemistry, 44, 2005
|
|
2NDH
 
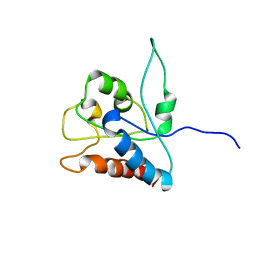 | | NMR solution structure of MAL/TIRAP TIR domain (C116A) | | Descriptor: | Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Lavrencic, P, Mobli, M. | | Deposit date: | 2016-05-27 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the TLR adaptor MAL/TIRAP reveals an intact BB loop and supports MAL Cys91 glutathionylation for signaling.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3GRR
 
 | |
5V0R
 
 | |
3E56
 
 | |
4HC4
 
 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein arginine N-methyltransferase 6, ... | | Authors: | Dong, A, Zeng, H, He, H, El Bakkouri, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of arginine asymmetrical dimethylation by PRMT6.
Biochem. J., 473, 2016
|
|
5W81
 
 | | Phosphorylated, ATP-bound structure of zebrafish cystic fibrosis transmembrane conductance regulator (CFTR) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Zhang, Z, Liu, F, Chen, J. | | Deposit date: | 2017-06-21 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Conformational Changes of CFTR upon Phosphorylation and ATP Binding.
Cell, 170, 2017
|
|
1NC3
 
 | | Crystal structure of E. coli MTA/AdoHcy nucleosidase complexed with formycin A (FMA) | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2002-12-04 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Escherichia coli 5'-methylthioadenosine/ S-adenosylhomocysteine nucleosidase inhibitor complexes provide insight into the conformational changes required for substrate binding and catalysis.
J.Biol.Chem., 278, 2003
|
|
4G67
 
 | |
3F8Z
 
 | | Human Dihydrofolate Reductase Structural Data with Active Site Mutant Enzyme Complexes | | Descriptor: | 2,4-DIAMINO-5-[2-METHOXY-5-(4-CARBOXYBUTYLOXY)BENZYL]PYRIMIDINE, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V, Pace, J, Makin, J, Piraino, J, Queener, S.F, Rosowsky, A. | | Deposit date: | 2008-11-13 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Correlations of Inhibitor Kinetics for Pneumocystis jirovecii and Human Dihydrofolate Reductase with Structural Data for Human Active Site Mutant Enzyme Complexes.
Biochemistry, 48, 2009
|
|
5N14
 
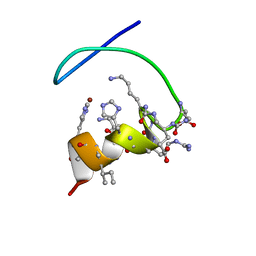 | |
2P1D
 
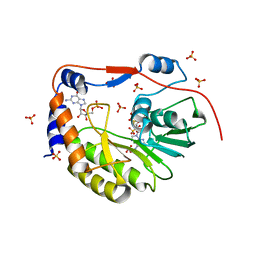 | |
3F91
 
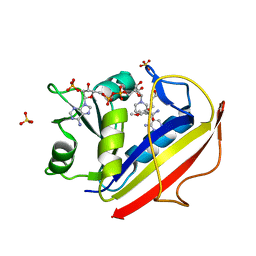 | | Structural Data for Human Active Site Mutant Enzyme Complexes | | Descriptor: | 2,4-DIAMINO-5-[2-METHOXY-5-(4-CARBOXYBUTYLOXY)BENZYL]PYRIMIDINE, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V, Pace, J, Makin, J, Piraino, J, Queener, S.F, Rosowsky, A. | | Deposit date: | 2008-11-13 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Correlations of Inhibitor Kinetics for Pneumocystis jirovecii and Human Dihydrofolate Reductase with Structural Data for Human Active Site Mutant Enzyme Complexes.
Biochemistry, 48, 2009
|
|
2ADM
 
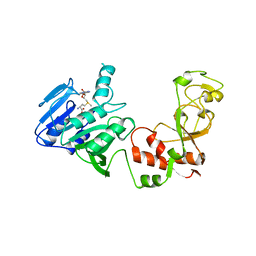 | | ADENINE-N6-DNA-METHYLTRANSFERASE TAQI | | Descriptor: | ADENINE-N6-DNA-METHYLTRANSFERASE TAQI, S-ADENOSYLMETHIONINE | | Authors: | Schluckebier, G, Saenger, W. | | Deposit date: | 1996-07-15 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Differential binding of S-adenosylmethionine S-adenosylhomocysteine and Sinefungin to the adenine-specific DNA methyltransferase M.TaqI.
J.Mol.Biol., 265, 1997
|
|
3F8Y
 
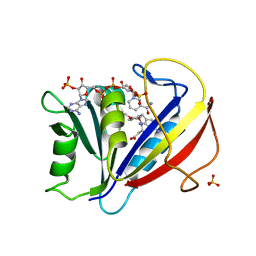 | | Correlations of Human Dihydrofolate Reductase with Structural Data for Human Active Site Mutant Enzyme Complexes | | Descriptor: | 2,4-DIAMINO-5-[2-METHOXY-5-(4-CARBOXYBUTYLOXY)BENZYL]PYRIMIDINE, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V, Pace, J, Makin, J, Piraino, J, Queener, S.F, Rosowsky, A. | | Deposit date: | 2008-11-13 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Correlations of Inhibitor Kinetics for Pneumocystis jirovecii and Human Dihydrofolate Reductase with Structural Data for Human Active Site Mutant Enzyme Complexes.
Biochemistry, 48, 2009
|
|
1NW7
 
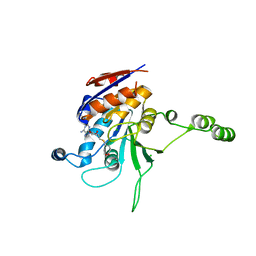 | | Structure of the beta class N6-adenine DNA methyltransferase RsrI bound to S-ADENOSYL-L-HOMOCYSTEINE | | Descriptor: | CHLORIDE ION, MODIFICATION METHYLASE RSRI, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Thomas, C.B, Scavetta, R.D, Gumport, R.I, Churchill, M.E.A. | | Deposit date: | 2003-02-05 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of liganded and unliganded RsrI N6-adenine DNA methyltransferase: a distinct orientation for active cofactor binding
J.Biol.Chem., 278, 2003
|
|
4KRG
 
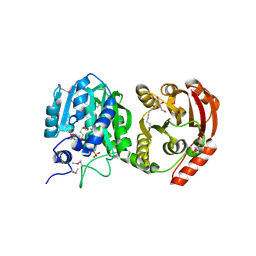 | |
4KIB
 
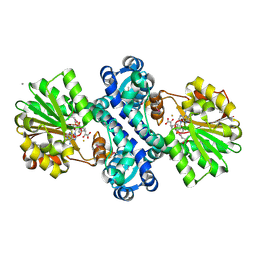 | | Crystal structure of methyltransferase from Streptomyces hygroscopicus complexed with S-adenosyl-L-homocysteine and methylphenylpyruvic acid | | Descriptor: | (2R)-2-hydroxy-3-phenylpropanoic acid, (3R)-2-oxo-3-phenylbutanoic acid, CALCIUM ION, ... | | Authors: | Liu, Y.C, Zou, X.W, Chan, H.C, Huang, C.J, Li, T.L. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of a nonhaem-iron SAM-dependent C-methyltransferase and its engineering to a hydratase and an O-methyltransferase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1JYS
 
 | | Crystal Structure of E. coli MTA/AdoHcy Nucleosidase | | Descriptor: | ADENINE, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2001-09-13 | | Release date: | 2002-10-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of E. coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase reveals similarity to the purine nucleoside phosphorylases.
Structure, 9, 2001
|
|
3NGV
 
 | | Crystal structure of AnSt-D7L1 | | Descriptor: | D7 protein, GLYCEROL, SULFATE ION | | Authors: | Andersen, J.F, Alvarenga, P.H, Francischetti, I.M, Ribeiro, J.M, Calvo, E. | | Deposit date: | 2010-06-13 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The function and three-dimensional structure of a thromboxane a(2)/cysteinyl leukotriene-binding protein from the saliva of a mosquito vector of the malaria parasite.
Plos Biol., 8, 2010
|
|
3NHI
 
 | | Crystal structure of the AnSt-D7L1-leukotriene C4 complex | | Descriptor: | (5S,7E,9E,11Z,14Z)-5-hydroxyicosa-7,9,11,14-tetraenoic acid, D7 protein, GLYCEROL, ... | | Authors: | Andersen, J.F, Alvarenga, P.H, Francischetti, I.M, Calvo, E, Sa-Nunes, A, Ribeiro, J.M. | | Deposit date: | 2010-06-14 | | Release date: | 2010-12-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The function and three-dimensional structure of a thromboxane a(2)/cysteinyl leukotriene-binding protein from the saliva of a mosquito vector of the malaria parasite.
Plos Biol., 8, 2010
|
|
3NHT
 
 | | Crystal structure of the AnSt-D7L1-U46619 complex | | Descriptor: | (5E)-7-{6-[(1E)-3-HYDROXYOCT-1-ENYL]-2-OXABICYCLO[2.2.1]HEPT-5-YL}HEPT-5-ENOIC ACID, D7 protein, SULFATE ION | | Authors: | Andersen, J.F, Alvarenga, P.H, Francischetti, I.M, Calvo, E, Sa-Nunes, A, Ribeiro, J.M. | | Deposit date: | 2010-06-14 | | Release date: | 2010-12-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The function and three-dimensional structure of a thromboxane a(2)/cysteinyl leukotriene-binding protein from the saliva of a mosquito vector of the malaria parasite.
Plos Biol., 8, 2010
|
|
3GWZ
 
 | | Structure of the Mitomycin 7-O-methyltransferase MmcR | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, MmcR, ... | | Authors: | Singh, S, Chang, A, Bingman, C.A, Phillips Jr, G.N, Thorson, J.S. | | Deposit date: | 2009-04-01 | | Release date: | 2010-04-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural characterization of the mitomycin 7-O-methyltransferase.
Proteins, 79, 2011
|
|
