1K5H
 
 | | 1-deoxy-D-xylulose-5-phosphate reductoisomerase | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate reductoisomerase | | Authors: | Reuter, K, Sanderbrand, S, Jomaa, H, Wiesner, J, Steinbrecher, I, Beck, E, Hintz, M, Klebe, G, Stubbs, M.T. | | Deposit date: | 2001-10-10 | | Release date: | 2002-02-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 1-deoxy-D-xylulose-5-phosphate reductoisomerase, a crucial enzyme in the non-mevalonate pathway of isoprenoid biosynthesis.
J.Biol.Chem., 277, 2002
|
|
8CNC
 
 | | Structure of compound 1 bound KMT9 | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl](3-aminopropyl)amino}-5'-deoxyadenosine, Methyltransferase N6AMT1, Multifunctional methyltransferase subunit TRM112-like protein | | Authors: | Sheng, W. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure of compound 1 bound KMT9
To Be Published
|
|
2IV1
 
 | |
7VSR
 
 | | Structure of McrBC (stalkless mutant) | | Descriptor: | 5-methylcytosine-specific restriction enzyme B, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Saikrishnan, K, Adhav, V.A, Bose, S, Kutti R, V. | | Deposit date: | 2021-10-27 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of McrBC (stalkless mutant)
To Be Published
|
|
1K9S
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE FROM E. COLI IN COMPLEX WITH FORMYCIN A DERIVATIVE AND PHOSPHATE | | Descriptor: | 2-(7-AMINO-6-METHYL-3H-PYRAZOLO[4,3-D]PYRIMIDIN-3-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, 2-HYDROXYMETHYL-5-(7-METHYLAMINO-3H-PYRAZOLO[4,3-D]PYRIMIDIN-3-YL)-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, ... | | Authors: | Koellner, G, Bzowska, A, Wielgus-Kutrowska, B, Luic, M, Steiner, T, Saenger, W, Stepinski, J. | | Deposit date: | 2001-10-30 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Open and closed conformation of the E. coli purine nucleoside phosphorylase active center and implications for the catalytic mechanism.
J.Mol.Biol., 315, 2002
|
|
1K44
 
 | |
1K5B
 
 | | Crystal Structure of Human Angiogenin Variant des(121-123) | | Descriptor: | Angiogenin, CITRIC ACID | | Authors: | Leonidas, D.D, Shapiro, R, Subbarao, G.V, Russo, A, Acharya, K.R. | | Deposit date: | 2001-10-10 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic studies on the role of the C-terminal segment of human angiogenin in defining enzymatic potency.
Biochemistry, 41, 2002
|
|
1K7H
 
 | | CRYSTAL STRUCTURE OF SHRIMP ALKALINE PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALKALINE PHOSPHATASE, MALEIC ACID, ... | | Authors: | De Backer, M.E, Mc Sweeney, S, Rasmussen, H.B, Riise, B.W, Lindley, P, Hough, E. | | Deposit date: | 2001-10-19 | | Release date: | 2002-07-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The 1.9 A Crystal Structure of Heat-Labile Shrimp Alkaline Phosphatase
J.Mol.Biol., 318, 2002
|
|
1K59
 
 | | Crystal Structure of Human Angiogenin Variant Q117G | | Descriptor: | CITRIC ACID, angiogenin | | Authors: | Leonidas, D.D, Shapiro, R, Subbarao, G.V, Russo, A, Acharya, K.R. | | Deposit date: | 2001-10-10 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic studies on the role of the C-terminal segment of human angiogenin in defining enzymatic potency.
Biochemistry, 41, 2002
|
|
2IWZ
 
 | | Human mitochondrial beta-ketoacyl ACP synthase complexed with hexanoic acid | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE, AMMONIUM ION, HEXANOIC ACID | | Authors: | Christensen, C.E, Kragelund, B.B, von Wettstein-Knowles, P, Henriksen, A. | | Deposit date: | 2006-07-05 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the Human Beta-Ketoacyl [Acp] Synthase from the Mitochondrial Type II Fatty Acid Synthase.
Protein Sci., 16, 2007
|
|
8C3C
 
 | |
1K7K
 
 | | crystal structure of RdgB- inosine triphosphate pyrophosphatase from E. coli | | Descriptor: | Hypothetical protein yggV | | Authors: | Sanishvili, R, Joachimiak, A, Edwards, A, Savchenko, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-19 | | Release date: | 2002-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of the antimutagenic activity of the house-cleaning inosine triphosphate pyrophosphatase RdgB from Escherichia coli.
J.Mol.Biol., 374, 2007
|
|
1K3F
 
 | | Uridine Phosphorylase from E. coli, Refined in the Monoclinic Crystal Lattice | | Descriptor: | uridine phosphorylase | | Authors: | Morgunova, E.Yu, Mikhailov, A.M, Popov, A.N, Blagova, E.V, Smirnova, E.A, Vainshtein, B.K, Mao, C, Armstrong, S.R, Ealick, S.E, Komissarov, A.A, Linkova, E.V, Burlakova, A.A, Mironov, A.S, Debabov, V.G. | | Deposit date: | 2001-10-02 | | Release date: | 2001-10-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic structure at 2.5 A resolution of uridine phosphorylase from E. coli as refined in the monoclinic crystal lattice.
FEBS Lett., 367, 1995
|
|
8C2G
 
 | |
1K5A
 
 | | Crystal structure of human angiogenin double variant I119A/F120A | | Descriptor: | Angiogenin | | Authors: | Leonidas, D.D, Shapiro, R, Subbarao, G.V, Russo, A, Acharya, K.R. | | Deposit date: | 2001-10-10 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystallographic studies on the role of the C-terminal segment of human angiogenin in defining enzymatic potency.
Biochemistry, 41, 2002
|
|
2IUO
 
 | | Site Directed Mutagenesis of Key Residues Involved in the Catalytic Mechanism of Cyanase | | Descriptor: | AZIDE ION, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Guilloton, M, Walsh, M.A, Joachimiak, A, Anderson, M.P. | | Deposit date: | 2006-06-06 | | Release date: | 2006-06-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Twin Set of Low Pka Arginines Ensures the Concerted Acid Base Catalytic Mechanism of Cyanase
To be Published
|
|
2IKF
 
 | |
1K5G
 
 | | Crystal structure of Ran-GDP-AlFx-RanBP1-RanGAP complex | | Descriptor: | ALUMINUM FLUORIDE, GTP-binding nuclear protein RAN, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Seewald, M.J, Koerner, C, Wittinghofer, A, Vetter, I.R. | | Deposit date: | 2001-10-10 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | RanGAP mediates GTP hydrolysis without an arginine finger.
Nature, 415, 2002
|
|
1K9T
 
 | | Chitinase a complexed with tetra-N-acetylchitotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Prag, G, Tucker, P.A, Oppenheim, A.B. | | Deposit date: | 2001-10-30 | | Release date: | 2002-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Complex Structures of Chitinase A Mutant with Oligonag Provide Insight Into the Enzymatic Mechanism
To be Published
|
|
2ITD
 
 | | Potassium Channel KcsA-Fab complex in Barium Chloride | | Descriptor: | BARIUM ION, Voltage-gated potassium channel, antibody Fab fragment heavy chain, ... | | Authors: | Lockless, S.W, Zhou, M, MacKinnon, R. | | Deposit date: | 2006-10-19 | | Release date: | 2007-05-15 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Thermodynamic Properties of Selective Ion Binding in a K(+) Channel.
Plos Biol., 5, 2007
|
|
5F28
 
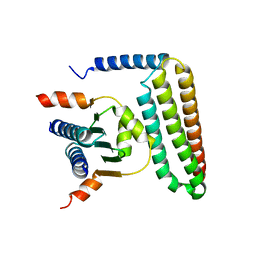 | | Crystal structure of FAT domain of Focal Adhesion Kinase (FAK) bound to the transcription factor MEF2C | | Descriptor: | Focal adhesion kinase 1, MEF2C | | Authors: | Cardoso, A.C, Ambrosio, A.L.B, Dessen, A, Franchini, K.G. | | Deposit date: | 2015-12-01 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | FAK Forms a Complex with MEF2 to Couple Biomechanical Signaling to Transcription in Cardiomyocytes.
Structure, 24, 2016
|
|
2ITC
 
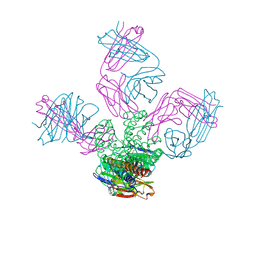 | | Potassium Channel KcsA-Fab complex in Sodium Chloride | | Descriptor: | Antibody Fab fragment heavy chain, Antibody Fab fragment light chain, SODIUM ION, ... | | Authors: | Lockless, S.W, Zhou, M, MacKinnon, R. | | Deposit date: | 2006-10-19 | | Release date: | 2007-05-15 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and Thermodynamic Properties of Selective Ion Binding in a K(+) Channel.
Plos Biol., 5, 2007
|
|
8BYO
 
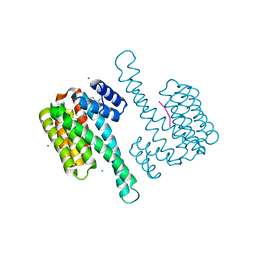 | | fragment-linked stabilizer for ERa - 14-3-3 interaction (1074361) | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, MAGNESIUM ION, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2023-01-18 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8C2D
 
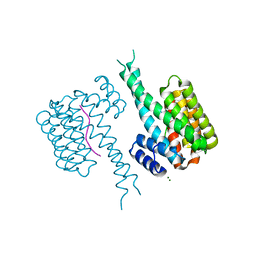 | | 14-3-3 in complex with Pyrin pS208 | | Descriptor: | 14-3-3 protein sigma, MAGNESIUM ION, Pyrin pS208 peptide | | Authors: | Lau, R, Ottmann, C, Hann, M. | | Deposit date: | 2022-12-22 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure and ligandability of the 14-3-3/pyrin interface.
Biochem.Biophys.Res.Commun., 651, 2023
|
|
8BZW
 
 | | Co-soaked stabilizers for ERa - 14-3-3 interaction (844_AZ210) | | Descriptor: | 14-3-3 protein sigma, 2-(4-chloranylphenoxy)-2-methyl-~{N}-(2-sulfanylethyl)propanamide, 4-[(2~{R})-3-azanyl-2-methyl-propyl]-7-methoxy-1-benzothiophene-2-carboximidamide, ... | | Authors: | Visser, E.J, Sijbesma, E, Ottmann, C. | | Deposit date: | 2022-12-15 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
