8QZ1
 
 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with a nanobody (Nb58) | | Descriptor: | Isoform B of Potassium channel subfamily K member 10, Nanobody 58, POTASSIUM ION | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Baronina, A, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.588 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
7BE7
 
 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
8RJY
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalent inhibitor GUE-3899 (compound 58 in publication) | | Descriptor: | 3C-like proteinase nsp5, ~{N}-[(2~{S})-1-[[(2~{S})-1-[[(4-chlorophenyl)methyl-(iminomethyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]thiophene-2-carboxamide | | Authors: | Strater, N, Claff, T, Sylvester, K, Mueller, C.E, Guetschow, M, Useini, A. | | Deposit date: | 2023-12-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Macrocyclic Azapeptide Nitriles: Structure-Based Discovery of Potent SARS-CoV-2 Main Protease Inhibitors as Antiviral Drugs.
J.Med.Chem., 67, 2024
|
|
7RT5
 
 | | Crystal structure of KRAS G12D with compound 36 (4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-7-(8-ethynyl-7-fluoronaphthalen-1-yl)-8-fluoro-2-{[(4s,7as)-tetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidine) bound | | Descriptor: | 4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-7-(8-ethynyl-7-fluoronaphthalen-1-yl)-8-fluoro-2-{[(4s,7as)-tetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidine, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-08-12 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
8RJZ
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GUE-3801 (compound 80 in publication) | | Descriptor: | (7~{S})-6-[2-[2,4-bis(chloranyl)phenoxy]ethanoyl]-14-fluoranyl-10-(iminomethyl)-9-methyl-7-(phenylmethyl)-2-oxa-6,9,10-triazabicyclo[10.4.0]hexadeca-1(12),13,15-trien-8-one, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase nsp5, ... | | Authors: | Strater, N, Claff, T, Sylvester, K, Mueller, C.E, Guetschow, M, Useini, A. | | Deposit date: | 2023-12-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Macrocyclic Azapeptide Nitriles: Structure-Based Discovery of Potent SARS-CoV-2 Main Protease Inhibitors as Antiviral Drugs.
J.Med.Chem., 67, 2024
|
|
6DLH
 
 | | Endo-fucoidan hydrolase MfFcnA4 from glycoside hydrolase family 107 | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-endofucoidanase, CALCIUM ION, ... | | Authors: | Vickers, C, Abe, K, Salama-Alber, O, Boraston, A.B. | | Deposit date: | 2018-06-01 | | Release date: | 2018-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to alpha-l-fucosidases from GH29.
J. Biol. Chem., 293, 2018
|
|
8B47
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a cyclic di-adenosine derivative | | Descriptor: | (1~{R},23~{R},24~{S},25~{R})-14-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-7-azanyl-24,25-bis(oxidanyl)-20,20-bis(oxidanylidene)-26-oxa-20$l^{6}-thia-2,4,6,9,14,17,21-heptazatetracyclo[21.2.1.0^{2,10}.0^{3,8}]hexacosa-3,5,7,9-tetraen-11-yn-16-one, NAD kinase 1, PENTAETHYLENE GLYCOL, ... | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2022-09-20 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Synthesis and structure-activity relationship studies of original cyclic diadenosine derivatives as nanomolar inhibitors of NAD kinase from pathogenic bacteria.
Eur.J.Med.Chem., 246, 2023
|
|
7C0L
 
 | |
8R9O
 
 | | A soakable crystal form of human CDK7 in complex with AMP-PNP | | Descriptor: | Cyclin-dependent kinase 7, ~{N}-[(1~{S})-2-(dimethylamino)-1-phenyl-ethyl]-6,6-dimethyl-3-[[4-(propanoylamino)phenyl]carbonylamino]-1,4-dihydropyrrolo[3,4-c]pyrazole-5-carboxamide | | Authors: | Mukherjee, M, Cleasby, A. | | Deposit date: | 2023-11-30 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Protein engineering enables a soakable crystal form of human CDK7 primed for high-throughput crystallography and structure-based drug design.
Structure, 32, 2024
|
|
7BBP
 
 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with H4K5acK8ac | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Histone H4, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with H4K5acK8ac
To Be Published
|
|
7RT3
 
 | | Crystal Structure of KRAS G12D with compound 24 (4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2S,4S,7aR)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol) bound | | Descriptor: | 4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2S,4S,7aR)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-08-12 | | Release date: | 2021-12-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
8RQH
 
 | | Crystal Structure of the flavoprotein monooxygenase TrlE from Streptomyces cyaneofuscatus Soc7 | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Sowa, S.T, Hoeing, L.S, Jakob, R.P, Maier, T, Teufel, R. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biosynthesis of the bacterial antibiotic 3,7-dihydroxytropolone through enzymatic salvaging of catabolic shunt products.
Chem Sci, 15, 2024
|
|
8R99
 
 | | A soakable crystal form of human CDK7 in complex with AMP-PNP | | Descriptor: | Cyclin-dependent kinase 7, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Mukherjee, M, Cleasby, A. | | Deposit date: | 2023-11-30 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Protein engineering enables a soakable crystal form of human CDK7 primed for high-throughput crystallography and structure-based drug design.
Structure, 32, 2024
|
|
6DN0
 
 | |
7BRS
 
 | | E.coli beta-galactosidase (E537Q) in complex with fluorescent probe KSA02 | | Descriptor: | 8-[2-[(E)-2-[4-[(2S,3R,4S,5R,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxyphenyl]ethenyl]-3,3-dimethyl-indol-1-ium-1-yl]octanoic acid, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Chen, X, Hu, Y.L, Gao, Y, Yuan, R, Guo, Y. | | Deposit date: | 2020-03-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Two-Dimensional Design Strategy to Construct Smart Fluorescent Probes for the Precise Tracking of Senescence.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8RND
 
 | | Cathepsin S in complex with NNPI-C10 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin S, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Petruzzella, A, Lau, K, Pojer, F, Oricchio, E. | | Deposit date: | 2024-01-09 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Antibody-peptide conjugates deliver covalent inhibitors blocking oncogenic cathepsins.
Nat.Chem.Biol., 20, 2024
|
|
7N6E
 
 | | TCR peptide HLA-A2 complex | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Spike protein S1, ... | | Authors: | Chaurasia, P, Rossjohn, J, Petersen, J. | | Deposit date: | 2021-06-08 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of biased T cell receptor recognition of an immunodominant HLA-A2 epitope of the SARS-CoV-2 spike protein.
J.Biol.Chem., 297, 2021
|
|
8BV0
 
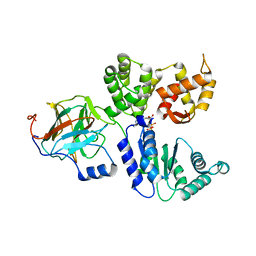 | | Binary complex between the NB-ARC domain from the Tomato immune receptor NRC1 and the SPRY domain-containing effector SS15 from the potato cyst nematode | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NRC1, Truncated secreted SPRY domain-containing protein 15 (Fragment) | | Authors: | Contreras, M.P, Pai, H, Muniyandi, S, Toghani, A, Lawson, D.M, Tumtas, Y, Duggan, C, Yuen, E.L.H, Stevenson, C.E.M, Harant, A, Wu, C.H, Bozkurt, T.O, Kamoun, S, Derevnina, L. | | Deposit date: | 2022-12-01 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Resurrection of plant disease resistance proteins via helper NLR bioengineering.
Sci Adv, 9, 2023
|
|
7BB2
 
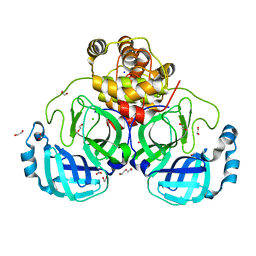 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 at 1.6A resolution (spacegroup P2(1)2(1)2(1)) | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2020-12-16 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
6IOS
 
 | | The ligand binding domain of Mlp24 with proline | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
7S6Q
 
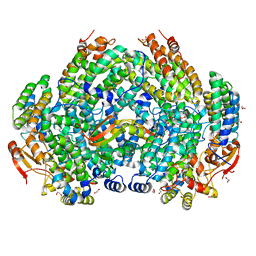 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit DBL2 | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, FE (III) ION, ... | | Authors: | Johns, J.C, Banerjee, R, Semonis, M.M, Shi, K, Aihara, H, Lipscomb, J.D. | | Deposit date: | 2021-09-14 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | X-ray Crystal Structures of Methane Monooxygenase Hydroxylase Complexes with Variants of Its Regulatory Component: Correlations with Altered Reaction Cycle Dynamics.
Biochemistry, 61, 2022
|
|
7C0Z
 
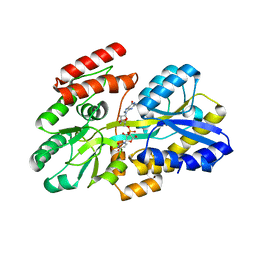 | | Crystal structure of a dinucleotide-binding protein (Y246A) of ABC transporter endogenously bound to uridylyl-3'-5'-phospho-guanosine (Form II) | | Descriptor: | 1,2-ETHANEDIOL, Sugar ABC transporter, periplasmic sugar-binding protein, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-01 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and thermodynamic insights into the novel dinucleotide-binding protein of ABC transporter unveils its moonlighting function.
Febs J., 288, 2021
|
|
7S86
 
 | |
8RDA
 
 | |
7N6D
 
 | | HLA peptide complex | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, MHC class I antigen, ... | | Authors: | Chaurasia, P, Petersen, J, Rossjohn, J. | | Deposit date: | 2021-06-08 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of biased T cell receptor recognition of an immunodominant HLA-A2 epitope of the SARS-CoV-2 spike protein.
J.Biol.Chem., 297, 2021
|
|
