1L26
 
 | |
1DQ9
 
 | | COMPLEX OF CATALYTIC PORTION OF HUMAN HMG-COA REDUCTASE WITH HMG-COA | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, PROTEIN (HMG-COA REDUCTASE) | | Authors: | Istvan, E.S, Palnitkar, M, Buchanan, S.K, Deisenhofer, J. | | Deposit date: | 1999-12-30 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis.
EMBO J., 19, 2000
|
|
1DRU
 
 | | ESCHERICHIA COLI DHPR/NADH COMPLEX | | Descriptor: | DIHYDRODIPICOLINATE REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Reddy, S.G, Scapin, G, Blanchard, J.S. | | Deposit date: | 1996-06-28 | | Release date: | 1997-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interaction of pyridine nucleotide substrates with Escherichia coli dihydrodipicolinate reductase: thermodynamic and structural analysis of binary complexes.
Biochemistry, 35, 1996
|
|
6I53
 
 | | Cryo-EM structure of the human synaptic alpha1-beta3-gamma2 GABAA receptor in complex with Megabody38 in a lipid nanodisc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Laverty, D, Desai, R, Uchanski, T, Masiulis, S, Wojciech, J.S, Malinauskas, T, Zivanov, J, Pardon, E, Steyaert, J, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-02 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the human alpha 1 beta 3 gamma 2 GABAAreceptor in a lipid bilayer.
Nature, 565, 2019
|
|
1L44
 
 | |
4DVY
 
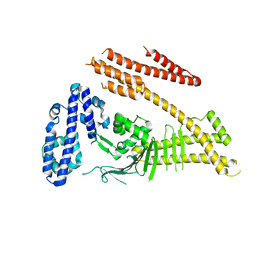 | | Crystal structure of the Helicobacter pylori CagA oncoprotein | | Descriptor: | Cytotoxicity-associated immunodominant antigen | | Authors: | Hayashi, T, Senda, M, Morohashi, H, Higashi, H, Horio, M, Kashiba, Y, Nagase, L, Sasaya, D, Shimizu, T, Venugopalan, N, Kumeta, H, Noda, N, Inagaki, F, Senda, T, Hatakeyama, M. | | Deposit date: | 2012-02-23 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Tertiary Structure-Function Analysis Reveals the Pathogenic Signaling Potentiation Mechanism of Helicobacter pylori Oncogenic Effector CagA
Cell Host Microbe, 12, 2012
|
|
1L58
 
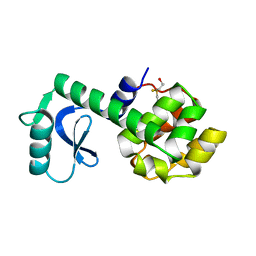 | |
1LJ3
 
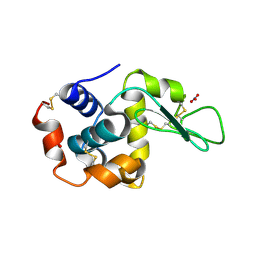 | |
1LJK
 
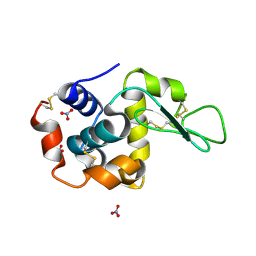 | |
1L64
 
 | |
1DYE
 
 | |
1LL7
 
 | |
3GHU
 
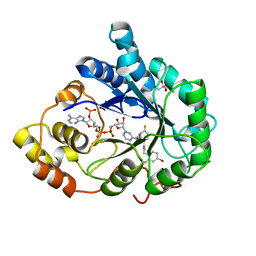 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594. Investigation of global effects of radiation damage on protein structure. Forth stage of radiation damage. | | Descriptor: | Aldose reductase, CITRIC ACID, IDD594, ... | | Authors: | Petrova, T, Ginell, S, Hazemann, I, Mitschler, A, Podjarny, A, Joachimiak, A. | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | X-ray-radiation-induced cooperative atomic movements in protein.
J.Mol.Biol., 387, 2009
|
|
1L71
 
 | |
4TM1
 
 | |
1PRN
 
 | |
4RQ5
 
 | |
3HHN
 
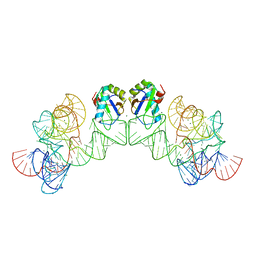 | | Crystal structure of class I ligase ribozyme self-ligation product, in complex with U1A RBD | | Descriptor: | Class I ligase ribozyme, self-ligation product, MAGNESIUM ION, ... | | Authors: | Shechner, D.M, Grant, R.A, Bagby, S.C, Bartel, D.P. | | Deposit date: | 2009-05-15 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.987 Å) | | Cite: | Crystal structure of the catalytic core of an RNA-polymerase ribozyme.
Science, 326, 2009
|
|
1L76
 
 | |
1DWH
 
 | |
1LQ2
 
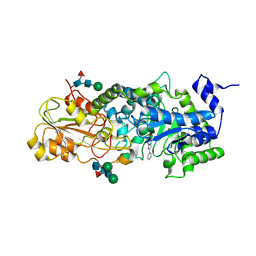 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with gluco-phenylimidazole | | Descriptor: | (5R,6R,7S,8S)-5-(HYDROXYMETHYL)-2-PHENYL-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hrmova, M, De Gori, R, Smith, B.J, Vasella, A, Varghese, J.N, Fincher, G.B. | | Deposit date: | 2002-05-09 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional Structure of the Barley {beta}-D-Glucan Glucohydrolase in Complex with a Transition State Mimic.
J.Biol.Chem., 279, 2004
|
|
6FZE
 
 | | BBE31 from Lyme disease agent Borrelia (Borreliella) burgdorferi playing a vital role in successful colonization of the mammalian host (native data) | | Descriptor: | GLUTATHIONE, Putative surface protein, SULFATE ION | | Authors: | Brangulis, K, Akopjana, I, Petrovskis, I, Kazaks, A, Tars, K. | | Deposit date: | 2018-03-14 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | BBE31 from the Lyme disease agent Borrelia burgdorferi, known to play an important role in successful colonization of the mammalian host, shows the ability to bind glutathione.
Biochim Biophys Acta Gen Subj, 1864, 2019
|
|
2Z4V
 
 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with magnesium and GGPP (inhibitory site) | | Descriptor: | GERANYLGERANYL DIPHOSPHATE, Geranylgeranyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Chen, C.K.-M, Guo, R.T, Hudock, M, Cao, R, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-06-26 | | Release date: | 2008-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Inhibition of geranylgeranyl diphosphate synthase by bisphosphonates: a crystallographic and computational investigation
J.Med.Chem., 51, 2008
|
|
2Z78
 
 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with BPH-806 | | Descriptor: | 3-(decyloxy)-5-(3,5-difluorophenyl)-1-(2,2-diphosphonoethyl)pyridinium, Geranylgeranyl pyrophosphate synthetase | | Authors: | Chen, C.K.-M, Guo, R.T, Hudock, M, Cao, R, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-08-16 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of geranylgeranyl diphosphate synthase by bisphosphonates: a crystallographic and computational investigation
J.Med.Chem., 51, 2008
|
|
1L91
 
 | |
