3WTV
 
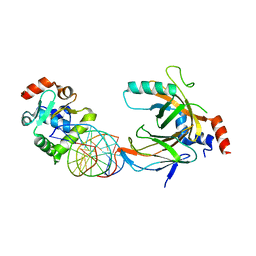 | | Crystal structure of the complex comprised of ETS1(V170G), RUNX1, CBFBETA, and the tcralpha gene enhancer DNA | | Descriptor: | Core-binding factor subunit beta, DNA (5'-D(*AP*GP*AP*GP*GP*AP*TP*GP*TP*GP*GP*CP*TP*TP*C)-3'), DNA (5'-D(*GP*AP*AP*GP*CP*CP*AP*CP*AP*TP*CP*CP*TP*CP*T)-3'), ... | | Authors: | Shiina, M, Hamada, K, Ogata, K. | | Deposit date: | 2014-04-21 | | Release date: | 2014-08-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A novel allosteric mechanism on protein-DNA interactions underlying the phosphorylation-dependent regulation of Ets1 target gene expressions.
J.Mol.Biol., 427, 2015
|
|
3ZSL
 
 | | Crystal structure of Apo Human Galectin-3 CRD at 1.08 angstrom resolution, at cryogenic temperature | | Descriptor: | GALECTIN-3 | | Authors: | Saraboji, K, Hakansson, M, Diehl, C, Nilsson, U.J, Leffler, H, Akke, M, Logan, D.T. | | Deposit date: | 2011-06-28 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | The Carbohydrate-Binding Site in Galectin-3 is Pre-Organized to Recognize a Sugar-Like Framework of Oxygens: Ultra-High Resolution Structures and Water Dynamics.
Biochemistry, 51, 2012
|
|
3ZKU
 
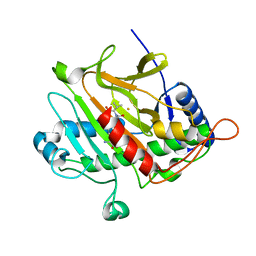 | | Isopenicillin N synthase with substrate analogue AhCV | | Descriptor: | FE (III) ION, ISOPENICILLIN N SYNTHASE, N-[(5S)-5-amino-5-carboxypentanoyl]-L-homocysteyl-D-valine | | Authors: | Daruzzaman, A, Clifton, I.J, Rutledge, P.J. | | Deposit date: | 2013-01-24 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Interaction of Isopenicillin N Synthase with Homologated Substrate Analogues Delta-(L-Alpha-Aminoadipoyl)-L-Homocysteinyl-D-Xaa Characterised by Protein Crystallography.
Chembiochem, 14, 2013
|
|
3ZGJ
 
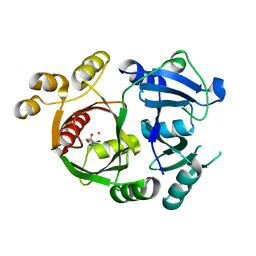 | |
3ZKY
 
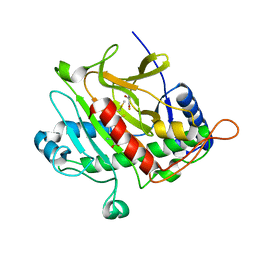 | | Isopenicillin N synthase with substrate analogue AhCmC | | Descriptor: | FE (III) ION, GLYCEROL, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Daruzzaman, A, Clifton, I.J, Rutledge, P.J. | | Deposit date: | 2013-01-25 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Interaction of Isopenicillin N Synthase with Homologated Substrate Analogues Delta-(L-Alpha-Aminoadipoyl)-L-Homocysteinyl-D-Xaa Characterised by Protein Crystallography.
Chembiochem, 14, 2013
|
|
3ZSM
 
 | | Crystal structure of Apo Human Galectin-3 CRD at 1.25 angstrom resolution, at room temperature | | Descriptor: | GALECTIN-3 | | Authors: | Saraboji, K, Hakansson, M, Diehl, C, Nilsson, U.J, Leffler, H, Akke, M, Logan, D.T. | | Deposit date: | 2011-06-28 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Carbohydrate-Binding Site in Galectin-3 is Pre-Organized to Recognize a Sugar-Like Framework of Oxygens: Ultra-High Resolution Structures and Water Dynamics.
Biochemistry, 51, 2012
|
|
3ZHB
 
 | |
3ZII
 
 | | Bacillus subtilis SepF G109K, C-terminal domain | | Descriptor: | CELL DIVISION PROTEIN SEPF | | Authors: | Duman, R, Ishikawa, S, Celik, I, Ogasawara, N, Lowe, J, Hamoen, L.W. | | Deposit date: | 2013-01-09 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Genetic Analyses Reveal the Protein Sepf as a New Membrane Anchor for the Z Ring.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4ACR
 
 | | Crystal structure of N-glycosylated, C-terminally truncated human glypican-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYPICAN-1 | | Authors: | Svensson, G, Awad, W, Mani, K, Logan, D.T. | | Deposit date: | 2011-12-17 | | Release date: | 2012-02-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of N-Glycosylated Human Glypican-1 Core Protein: Structure of Two Loops Evolutionarily Conserved in Vertebrate Glypican-1.
J.Biol.Chem., 287, 2012
|
|
3X0W
 
 | | Crystal structure of PLEKHM1 LIR-fused human LC3B_2-119 | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Suzuki, H, McEwan, D.G, Popovic, D, Gubas, A, Terawaki, S, Stadel, D, Coxon, F, Stegmann, D.M, Bhogaraju, S, Maddi, K, Kirchhoff, A, Gatti, E, Helfrich, M.H, Behrends, C, Pierre, P, Dikic, I, Wakatsuki, S. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins.
Mol.Cell, 57, 2015
|
|
3ZDN
 
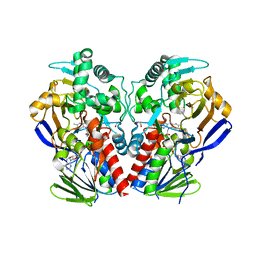 | | D11-C mutant of monoamine oxidase from Aspergillus niger | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, MONOAMINE OXIDASE N | | Authors: | Frank, A, Ghislieri, D, Willies, S, Turner, N.J, Grogan, G. | | Deposit date: | 2012-11-29 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Engineering an Enantioselective Amine Oxidase for the Synthesis of Pharmaceutical Building Blocks and Alkaloid Natural Products.
J.Am.Chem.Soc., 135, 2013
|
|
3ZIH
 
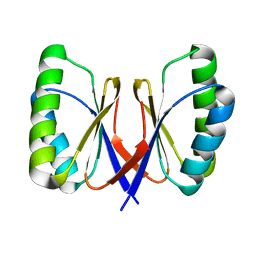 | | Bacillus subtilis SepF, C-terminal domain | | Descriptor: | CELL DIVISION PROTEIN SEPF | | Authors: | Duman, R.E, Ishikawa, S, Celik, I, Ogasawara, N, Lowe, J, Hamoen, L.W. | | Deposit date: | 2013-01-09 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Genetic Analyses Reveal the Protein Sepf as a New Membrane Anchor for the Z Ring.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ZGQ
 
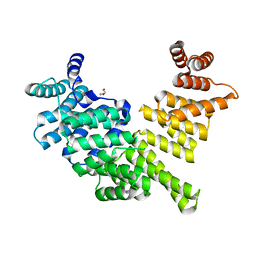 | | Crystal structure of human interferon-induced protein IFIT5 | | Descriptor: | DI(HYDROXYETHYL)ETHER, INTERFERON-INDUCED PROTEIN WITH TETRATRICOPEPTIDE REPEATS 5 | | Authors: | Katibah, G.E, Lee, H.J, Huizar, J.P, Vogan, J.M, Alber, T, Collins, K. | | Deposit date: | 2012-12-19 | | Release date: | 2013-01-23 | | Last modified: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | TRNA Binding, Structure, and Localization of the Human Interferon-Induced Protein Ifit5.
Mol.Cell, 49, 2013
|
|
4NHU
 
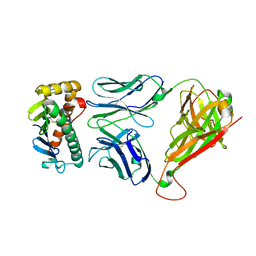 | | The M33 TCR p3M33l/H-2 Ld Complex | | Descriptor: | 2C m33 alpha VmCh chimera, 2C m33 beta VmCh chimera, H-2 class I histocompatibility antigen, ... | | Authors: | Birnbaum, M.E, Adams, J.J, Garcia, K.C. | | Deposit date: | 2013-11-05 | | Release date: | 2015-05-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Interrogating TCR Signal Strength and Cross-Reactivity by Yeast Display of pMHC
To be Published
|
|
3V10
 
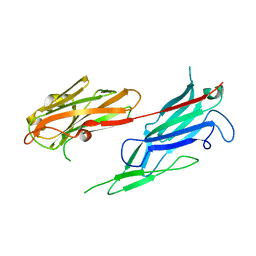 | | Crystal structure of the collagen binding domain of Erysipelothrix rhusiopathiae surface protein RspB | | Descriptor: | Rhusiopathiae surface protein B | | Authors: | Ponnuraj, K, Swarmistha devi, A, Ogawa, Y, Shimoji, Y, Subramainan, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-10-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Collagen adhesin-nanoparticle interaction impairs adhesin's ligand binding mechanism
Biochim.Biophys.Acta, 1820, 2012
|
|
4AC5
 
 | | Lipidic sponge phase crystal structure of the Bl. viridis reaction centre solved using serial femtosecond crystallography | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Johansson, L.C, Arnlund, D, White, T.A, Katona, G, DePonte, D.P, Weierstall, U, Doak, R.B, Shoeman, R.L, Lomb, L, Malmerberg, E, Davidsson, J, Nass, K, Liang, M, Andreasson, J, Aquila, A, Bajt, S, Barthelmess, M, Barty, A, Bogan, M.J, Bostedt, C, Bozek, J.D, Caleman, C, Coffee, R, Coppola, N, Ekeberg, T, Epp, S.W, Erk, B, Fleckenstein, H, Foucar, L, Graafsma, H, Gumprecht, L, Hajdu, J, Hampton, C.Y, Hartmann, R, Hartmann, A, Hauser, G, Hirsemann, H, Holl, P, Hunter, M.S, Kassemeyer, S, Kimmel, N, Kirian, R.A, Maia, F.R.N.C, Marchesini, S, Martin, A.V, Reich, C, Rolles, D, Rudek, B, Rudenko, A, Schlichting, I, Schulz, J, Seibert, M.M, Sierra, R, Soltau, H, Starodub, D, Stellato, F, Stern, S, Struder, L, Timneanu, N, Ullrich, J, Wahlgren, W.Y, Wang, X, Weidenspointner, G, Wunderer, C, Fromme, P, Chapman, H.N, Spence, J.C.H, Neutze, R. | | Deposit date: | 2011-12-14 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (8.2 Å) | | Cite: | Lipidic Phase Membrane Protein Serial Femtosecond Crystallography.
Nat.Methods, 9, 2012
|
|
4AD7
 
 | | Crystal structure of full-length N-glycosylated human glypican-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYPICAN-1 | | Authors: | Svensson, G, Awad, W, Mani, K, Logan, D.T. | | Deposit date: | 2011-12-22 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.945 Å) | | Cite: | Crystal Structure of N-Glycosylated Human Glypican-1 Core Protein: Structure of Two Loops Evolutionarily Conserved in Vertebrate Glypican-1.
J.Biol.Chem., 287, 2012
|
|
3W20
 
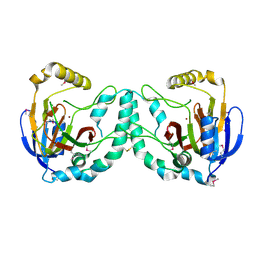 | | Crystal Structure of a Novel N-Substituted L-Amino Acid Dioxygenase from Burkholderia ambifaria AMMD | | Descriptor: | Putative uncharacterized protein, ZINC ION | | Authors: | Qin, H.M, Miyakawa, T, Jia, M.Z, Nakamura, A, Ohtsuka, J, Xue, Y.L, Kawashima, T, Kasahara, T, Hibi, M, Ogawa, J, Tanokura, M. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of a Novel N-Substituted L-Amino Acid Dioxygenase from Burkholderia ambifaria AMMD
Plos One, 8, 2013
|
|
3VJ7
 
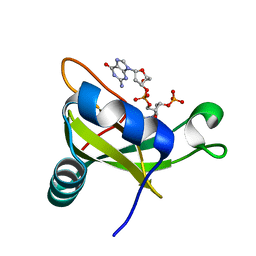 | | Crystal structure of the carboxy-terminal ribonuclease domain of Colicin E5 R33Q mutant | | Descriptor: | 2'-DEOXYURIDINE 3'-MONOPHOSPHATE, 2-AMINO-9-(2-DEOXY-3-O-PHOSPHONOPENTOFURANOSYL)-1,9-DIHYDRO-6H-PURIN-6-ONE, Colicin-E5 | | Authors: | Yajima, S, Inoue, S, Fushinobu, S, Ogawa, T, Hidaka, M, Masaki, H. | | Deposit date: | 2011-10-13 | | Release date: | 2011-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of the catalytic residues of sequence-specific and histidine-free ribonuclease colicin E5
J.Biochem., 152, 2012
|
|
4A6G
 
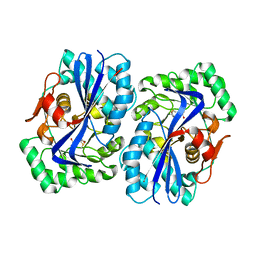 | | N-acyl amino acid racemase from Amycalotopsis sp. Ts-1-60: G291D- F323Y mutant in complex with N-acetyl methionine | | Descriptor: | MAGNESIUM ION, N-ACETYLMETHIONINE, N-ACYLAMINO ACID RACEMASE | | Authors: | Baxter, S, Royer, S, Grogan, G, Holt-Tiffin, K.E, Taylor, I.N, Fotheringham, I.G, Campopiano, D.J. | | Deposit date: | 2011-11-02 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | An Improved Racemase/Acylase Biotransformation for the Preparation of Enantiomerically Pure Amino Acids.
J.Am.Chem.Soc., 134, 2012
|
|
3ZZP
 
 | | Circular permutant of ribosomal protein S6, lacking edge strand beta- 2 of wild-type S6. | | Descriptor: | RIBOSOMAL PROTEIN S6 | | Authors: | Saraboji, K, Haglund, E, Lindberg, M.O, Oliveberg, M, Logan, D.T. | | Deposit date: | 2011-09-02 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Trimming Down a Protein Structure to its Bare Foldons: Spatial Organization of the Cooperative Unit.
J.Biol.Chem., 287, 2012
|
|
3ZIG
 
 | | SepF-like protein from Pyrococcus furiosus | | Descriptor: | SEPF-LIKE PROTEIN | | Authors: | Duman, R, Ishikawa, S, Celik, I, Ogasawara, N, Lowe, J, Hamoen, L.W. | | Deposit date: | 2013-01-09 | | Release date: | 2013-11-27 | | Last modified: | 2013-12-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Genetic Analyses Reveal the Protein Sepf as a New Membrane Anchor for the Z Ring.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ZIE
 
 | | SepF-like protein from Archaeoglobus fulgidus | | Descriptor: | SEPF-LIKE PROTEIN | | Authors: | Duman, R, Ishikawa, S, Celik, I, Ogasawara, N, Lowe, J, Hamoen, L.W. | | Deposit date: | 2013-01-08 | | Release date: | 2013-11-20 | | Last modified: | 2013-12-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Genetic Analyses Reveal the Protein Sepf as a New Membrane Anchor for the Z Ring
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3VG3
 
 | | Cadmium derivative of human LFABP | | Descriptor: | CADMIUM ION, Fatty acid-binding protein, liver, ... | | Authors: | Sharma, A, Yogavel, M, Sharma, A. | | Deposit date: | 2011-08-03 | | Release date: | 2012-06-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Utility of anion and cation combinations for phasing of protein structures.
J.Struct.Funct.Genom., 13, 2012
|
|
4ALB
 
 | | Structure of Phenolic Acid Decarboxylase from Bacillus subtilis: Tyr19Ala mutant in complex with coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHENOLIC ACID DECARBOXYLASE PADC | | Authors: | Frank, A, Eborall, W, Hyde, R, Hart, S, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2012-03-02 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Mutational Analysis of Phenolic Acid Decarboxylase from Bacillus Subtilis (Bspad), which Converts Bio-Derived Phenolic Acids to Styrene Derivatives
Catal.Sci.Technol., 2, 2012
|
|
