5AFJ
 
 | | alpha7-AChBP in complex with lobeline and fragment 1 | | Descriptor: | (3S)-6-(4-bromophenyl)-3-hydroxy-1,3-dimethyl-2,3-dihydropyridin-4(1H)-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE-BINDING PROTEIN, ... | | Authors: | Spurny, R, Debaveye, S, Farinha, A, Veys, K, Gossas, T, Atack, J, Bertrand, D, Kemp, J, Vos, A, Danielson, U.H, Tresadern, G, Ulens, C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Blueprint of Allosteric Binding Sites in a Homologue of the Agonist-Binding Domain of the Alpha7 Nicotinic Acetylcholine Receptor.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3K3G
 
 | | Crystal Structure of the Urea Transporter from Desulfovibrio Vulgaris Bound to 1,3-dimethylurea | | Descriptor: | 1,3-dimethylurea, GOLD ION, Urea transporter | | Authors: | Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2009-10-02 | | Release date: | 2009-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a bacterial homologue of the kidney urea transporter.
Nature, 462, 2009
|
|
7FJK
 
 | |
6V8K
 
 | |
5KZW
 
 | | Crystal structure of human GAA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Deming, D.T, Garman, S.C. | | Deposit date: | 2016-07-25 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of human GAA: structural basis of Pompe disease
To be published
|
|
5ZLJ
 
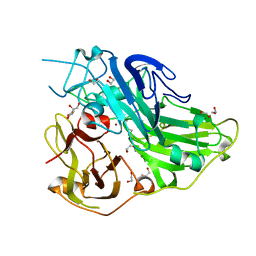 | | Crystal structure of CotA native enzyme, PH8.0 | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Xie, T, Liu, Z.C, Wang, G.G. | | Deposit date: | 2018-03-28 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Insight into the Allosteric Coupling of Cu1 Site and Trinuclear Cu Cluster in CotA Laccase.
Chembiochem, 19, 2018
|
|
6VCX
 
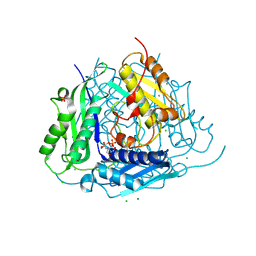 | | Crystal structure of Arabidopsis thaliana S-adenosylmethionine Synthase 1 (AtMAT1) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
9FBY
 
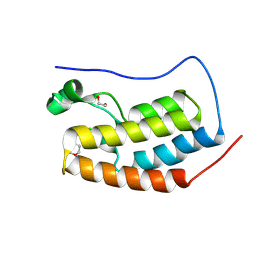 | |
9FBX
 
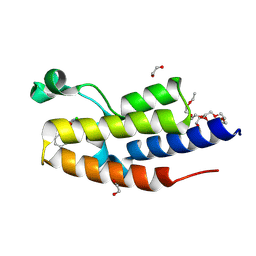 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 5-(1-benzyl-4-chloro-1H-imidazol-2-yl)-1,3-dimethylpyridin-2(1H)-one | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 5-(1-benzyl-4-chloro-1H-imidazol-2-yl)-1,3-dimethylpyridin-2(1H)-one, ... | | Authors: | Chung, C. | | Deposit date: | 2024-05-14 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Structure- and Property-Based Optimization of Efficient Pan-Bromodomain and Extra Terminal Inhibitors to Identify Oral and Intravenous Candidate I-BET787.
J.Med.Chem., 67, 2024
|
|
8PK0
 
 | | human mitoribosomal large subunit assembly intermediate 1 with GTPBP10-GTPBP7 | | Descriptor: | 16S rRNA + pre-H68-71 segment, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Kummer, E, Nguyen, T.G, Ritter, C. | | Deposit date: | 2023-06-23 | | Release date: | 2023-12-13 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insights into the role of GTPBP10 in the RNA maturation of the mitoribosome.
Nat Commun, 14, 2023
|
|
6Q2A
 
 | |
9EPP
 
 | | Cryo-EM Structure of Jumping Spider Rhodopsin-1 bound to a Giq heterotrimer | | Descriptor: | 11,20-Ethanoretinal, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tejero, O, Pamula, F, Koyanagi, M, Nagata, T, Afanasyev, P, Das, I, Deupi, X, Sheves, M, Terakita, A, Schertler, G.F.X, Rodrigues, M.J, Tsai, C.-J. | | Deposit date: | 2024-03-19 | | Release date: | 2024-10-23 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Active state structures of a bistable visual opsin bound to G proteins.
Nat Commun, 15, 2024
|
|
9FE0
 
 | | Crystal Structure of reduced NuoEF variant R66G(NuoF) from Aquifex aeolicus bound to NAD+ | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9EPQ
 
 | | Cryo-EM Structure of Jumping Spider Rhodopsin-1 bound to a Giq heterotrimer | | Descriptor: | 11,20-Ethanoretinal, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tejero, O, Pamula, F, Koyanagi, M, Nagata, T, Afanasyev, P, Das, I, Deupi, X, Sheves, M, Terakita, A, Schertler, G.F.X, Rodrigues, M.J, Tsai, C.-J. | | Deposit date: | 2024-03-19 | | Release date: | 2024-10-23 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Active state structures of a bistable visual opsin bound to G proteins.
Nat Commun, 15, 2024
|
|
9ETQ
 
 | | Crystal structure of PARP1 catalytic domain bound to AZD5305 (SARUPARIB) | | Descriptor: | 5-[4-[(7-ethyl-6-oxidanylidene-5~{H}-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl]-~{N}-methyl-pyridine-2-carboxamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M. | | Deposit date: | 2024-03-26 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery of 6-Fluoro-5-{4-[(5-fluoro-2-methyl-3-oxo-3,4-dihydroquinoxalin-6-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD9574): A CNS-Penetrant, PARP1-Selective Inhibitor.
J.Med.Chem., 67, 2024
|
|
8PQI
 
 | | PDGFRA T674I mutant kinase domain in complex with avapritinib derivative 9 | | Descriptor: | (1~{S})-~{N}-ethyl-1-(4-fluorophenyl)-1-[2-[4-[6-(1-methylpyrazol-4-yl)pyrrolo[2,1-f][1,2,4]triazin-4-yl]piperazin-1-yl]pyrimidin-5-yl]ethanamine, DI(HYDROXYETHYL)ETHER, Platelet-derived growth factor receptor alpha | | Authors: | Teuber, A, Kleinboelting, S, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-07-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Avapritinib-based SAR studies unveil a binding pocket in KIT and PDGFRA.
Nat Commun, 15, 2024
|
|
9FIF
 
 | | Crystal Structure of NuoEF variant P228R(NuoF) from Aquifex aeolicus bound to NADH under anoxic conditions | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-29 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
6GVH
 
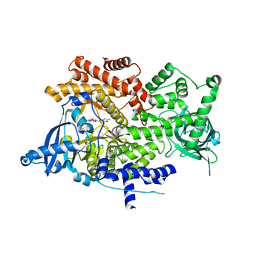 | | Crystal structure of PI3K alpha in complex with 3-(2-Amino-benzooxazol-5-yl)-4-chloro-1-isopropyl-1H-pyrazolo[3,4-d]pyrimidin-6-ylamine | | Descriptor: | 5-(6-azanyl-4-chloranyl-1-propan-2-yl-pyrazolo[3,4-d]pyrimidin-3-yl)-1,3-benzoxazol-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Ouvry, G, Aurelly, M, Bonnary, L, Borde, E, Bouix-Peter, C, Chantalat, L, Clary, L, Defoin-Platel, C, Deret, S, Forissier, M, Harris, C.S, Isabet, T, Lamy, L, Luzy, A.P, Pascau, J, Soulet, C, Taddei, A, Taquet, N, Tomas, L, Thoreau, E, Varvier, E, Vial, E, Hennequin, L.F. | | Deposit date: | 2018-06-21 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Impact of Minor Structural Modifications on Properties of a Series of mTOR Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
5A64
 
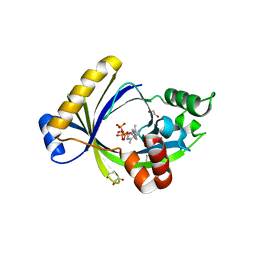 | | Crystal structure of mouse thiamine triphosphatase in complex with thiamine triphosphate. | | Descriptor: | 1,2-ETHANEDIOL, THIAMINE TRIPHOSPHATASE, TRIETHYLENE GLYCOL, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
5C08
 
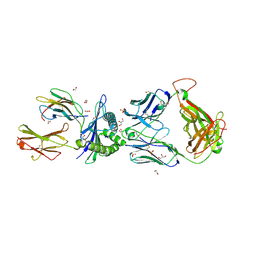 | | 1E6 TCR in Complex with HLA-A0e carrying RQWGPDPAAV | | Descriptor: | 1,2-ETHANEDIOL, 1E6 TCR Alpha Chain, 1E6 TCR Beta Chain, ... | | Authors: | Rizkallah, P.J, Bulek, A.M, Cole, D.K, Sewell, A.K. | | Deposit date: | 2015-06-12 | | Release date: | 2016-05-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.332 Å) | | Cite: | Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity.
J.Clin.Invest., 126, 2016
|
|
5NCY
 
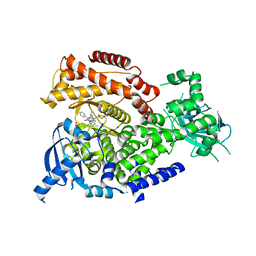 | | mPI3Kd IN COMPLEX WITH inh1 | | Descriptor: | 1,2-ETHANEDIOL, 4-azanyl-6-[[(1~{S})-1-(3-methyl-5-oxidanylidene-6-phenyl-[1,3]thiazolo[3,2-a]pyridin-7-yl)ethyl]amino]pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ... | | Authors: | Petersen, J. | | Deposit date: | 2017-03-06 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and Synthesis of Soluble and Cell-Permeable PI3K delta Inhibitors for Long-Acting Inhaled Administration.
J. Med. Chem., 60, 2017
|
|
5NHT
 
 | | human 199.54-16 TCR in complex with Melan-A/MART-1 (26-35) peptide and HLA-A2 | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, HLA class I histocompatibility antigen, ... | | Authors: | Exertier, C, Reiser, J.-B, Lantez, V, Chouquet, A, Bonneville, M, Saulquin, X, Housset, D. | | Deposit date: | 2017-03-22 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | human 199.54-16 TCR in complex with Melan-A/MART-1 (26-35) peptide and HLA-A2
To Be Published
|
|
8IC5
 
 | | Respiratory complex CIII2, focus-refined of type I, PERK -/- mouse under cold temperature | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, CARDIOLIPIN, ... | | Authors: | Shin, Y.-C, Liao, M. | | Deposit date: | 2023-02-10 | | Release date: | 2024-09-18 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of respiratory complex adaptation to cold temperatures.
Cell, 187, 2024
|
|
5URQ
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor p176 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-{2-chloro-5-[({2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}carbamoyl)amino]phenyl}-alpha-D-ribofuranosylamine, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-12 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor p176
To Be Published
|
|
6GBF
 
 | |
