7ASD
 
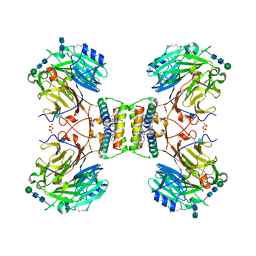 | | Structure of native royal jelly filaments | | Descriptor: | (3beta,14beta,17alpha)-ergosta-5,24(28)-dien-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mattei, S, Ban, A, Picenoni, A, Leibundgut, M, Glockshuber, R, Boehringer, D. | | Deposit date: | 2020-10-27 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of native glycolipoprotein filaments in honeybee royal jelly.
Nat Commun, 11, 2020
|
|
1V5C
 
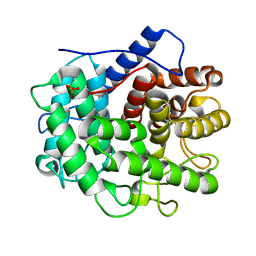 | | The crystal structure of the inactive form chitosanase from Bacillus sp. K17 at pH3.7 | | Descriptor: | SULFATE ION, chitosanase | | Authors: | Adachi, W, Shimizu, S, Sunami, T, Fukazawa, T, Suzuki, M, Yatsunami, R, Nakamura, S, Takenaka, A. | | Deposit date: | 2003-11-22 | | Release date: | 2004-12-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of family GH-8 chitosanase with subclass II specificity from Bacillus sp. K17
J.MOL.BIOL., 343, 2004
|
|
3FL7
 
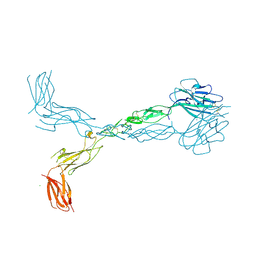 | | Crystal structure of the human ephrin A2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Ephrin receptor, ... | | Authors: | Walker, J.R, Yermekbayeva, L, Seitova, A, Butler-Cole, C, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-18 | | Release date: | 2009-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architecture of Eph receptor clusters.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
197L
 
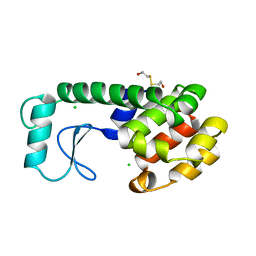 | | THERMODYNAMIC AND STRUCTURAL COMPENSATION IN "SIZE-SWITCH" CORE-REPACKING VARIANTS OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Baldwin, E, Xu, J, Hajiseyedjavadi, O, Matthews, B.W. | | Deposit date: | 1995-11-06 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thermodynamic and structural compensation in "size-switch" core repacking variants of bacteriophage T4 lysozyme.
J.Mol.Biol., 259, 1996
|
|
4IZT
 
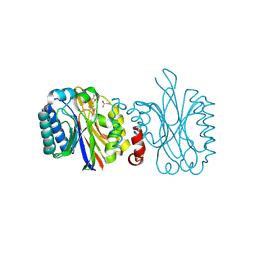 | |
147L
 
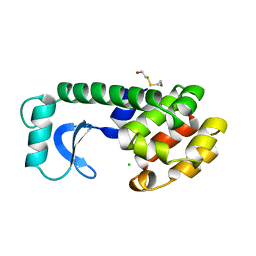 | |
7AJH
 
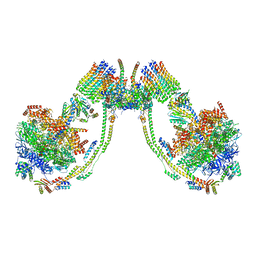 | | bovine ATP synthase dimer state3:state1 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ATP synthase F(0) complex subunit B1, mitochondrial, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-03 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Interface mobility between monomers in dimeric bovine ATP synthase participates in the ultrastructure of inner mitochondrial membranes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
163L
 
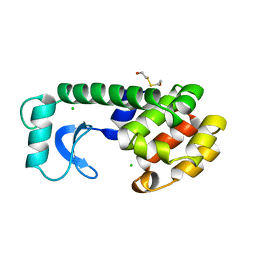 | | CONTROL OF ENZYME ACTIVITY BY AN ENGINEERED DISULFIDE BOND | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Blaber, M, Matthews, B.W. | | Deposit date: | 1994-06-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alanine scanning mutagenesis of the alpha-helix 115-123 of phage T4 lysozyme: effects on structure, stability and the binding of solvent.
J.Mol.Biol., 246, 1995
|
|
3FM0
 
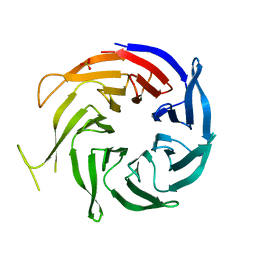 | | Crystal structure of WD40 protein Ciao1 | | Descriptor: | Protein CIAO1, SULFATE ION | | Authors: | Dong, A, Ravichandran, M, Crombet, L, Cossar, D, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-19 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
3P6Q
 
 | |
5ABU
 
 | |
182L
 
 | |
4J1B
 
 | |
3OVM
 
 | | X-ray Structural study of quinone reductase II inhibition by compounds with micromolar to nanomolar range IC50 values | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ZINC ION, ... | | Authors: | Pegan, S.D, Sturdy, M, Ferry, G, Delagrange, P, Boutin, J.A. | | Deposit date: | 2010-09-16 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | X-ray structural studies of quinone reductase 2 nanomolar range inhibitors.
Protein Sci., 20, 2011
|
|
7AJD
 
 | | bovine ATP synthase dimer state1:state3 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ATP synthase F(0) complex subunit B1, mitochondrial, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-03 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Interface mobility between monomers in dimeric bovine ATP synthase participates in the ultrastructure of inner mitochondrial membranes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4J28
 
 | | Crystal structure of a gh29 alpha-l-fucosidase gh29 from bacteroides thetaiotaomicron in complex with a 5-membered iminocyclitol inhibitor | | Descriptor: | (2S,3S,4R,5S)-2-(1H-benzimidazol-2-yl)-5-methylpyrrolidine-3,4-diol, Alpha-L-fucosidase, GLYCEROL, ... | | Authors: | Wright, D.W. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Three dimensional structure of a bacterial alpha-l-fucosidase with a 5-membered iminocyclitol inhibitor.
Bioorg.Med.Chem., 21, 2013
|
|
4J2M
 
 | |
3S81
 
 | | Crystal Structure of Putative Aspartate Racemase from Salmonella Typhimurium | | Descriptor: | CHLORIDE ION, Putative aspartate racemase, SULFATE ION | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Zhang, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-27 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Crystal Structure of Putative Aspartate Racemase from Salmonella Typhimurium
To be Published
|
|
1UKR
 
 | | STRUCTURE OF ENDO-1,4-BETA-XYLANASE C | | Descriptor: | ENDO-1,4-B-XYLANASE I | | Authors: | Krengel, U, Dijkstra, B.W. | | Deposit date: | 1996-08-23 | | Release date: | 1997-12-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-dimensional structure of Endo-1,4-beta-xylanase I from Aspergillus niger: molecular basis for its low pH optimum.
J.Mol.Biol., 263, 1996
|
|
2VEX
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B IN COMPLEX WITH AN ISOTHIAZOLIDINONE-CONTAINING INHIBITOR | | Descriptor: | MAGNESIUM ION, N-{(1S)-2-{4-[(5S)-1,1-dioxido-3-oxoisothiazolidin-5-yl]phenyl}-1-[(4R)-4-(2-phenylethyl)-4,5-dihydro-1H-imidazol-2-yl]ethyl}-3-fluorobenzenesulfonamide, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Douty, B, Wayland, B, Ala, P.J, Bower, M.J, Pruitt, J, Bostrom, L, Wei, M, Klabe, R, Gonneville, L, Wynn, R, Burn, T.C, Liu, P.C.C, Combs, A.P, Yue, E.W. | | Deposit date: | 2007-10-27 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isothiazolidinone Inhibitors of Ptp1B Containing Imidazoles and Imidazolines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1UKW
 
 | | Crystal structure of medium-chain acyl-CoA dehydrogenase from Thermus thermophilus HB8 | | Descriptor: | COBALT (II) ION, FLAVIN-ADENINE DINUCLEOTIDE, acyl-CoA dehydrogenase | | Authors: | Hamada, K, Ago, H, Kuramitsu, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-02 | | Release date: | 2004-11-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Thermus thermophilus medium-chain acyl-CoA dehydrogenase
To be published
|
|
7AJC
 
 | | bovine ATP synthase dimer state1:state2 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ATP synthase F(0) complex subunit B1, mitochondrial, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-03 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (11.9 Å) | | Cite: | Interface mobility between monomers in dimeric bovine ATP synthase participates in the ultrastructure of inner mitochondrial membranes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3SLE
 
 | |
3SAU
 
 | | MUTM Interrogation complex 6 | | Descriptor: | 5'-D(*A*GP*GP*TP*AP*GP*AP*CP*CP*AP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*T*CP*CP*TP*GP*GP*(TX2) P*CP*TP*AP*CP*C)-3', DNA GLYCOSYLASE, ... | | Authors: | Spong, M.C, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-06-03 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Strandwise translocation of a DNA glycosylase on undamaged DNA.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5AAG
 
 | | Aurora A kinase bound to an imidazopyridine inhibitor (14b) | | Descriptor: | AURORA KINASE A, [3-[[4-[6-chloranyl-2-(1,3-dimethylpyrazol-4-yl)-3H-imidazo[4,5-b]pyridin-7-yl]pyrazol-1-yl]methyl]phenyl]-(4-methylpiperazin-1-yl)methanone | | Authors: | McIntyre, P.J, Bayliss, R. | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | 7-(Pyrazol-4-Yl)-3H-Imidazo[4,5-B]Pyridine-Based Derivatives for Kinase Inhibition: Co-Crystallisation Studies with Aurora-A Reveal Distinct Differences in the Orientation of the Pyrazole N1-Substituent.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
