5J8K
 
 | | Architecture of supercomplex I-III2 | | Descriptor: | COMPLEX I 13KDA/NDUFS6, COMPLEX I 15KDA/NDUFS5, COMPLEX I 18KDA/NDUFS6, ... | | Authors: | Letts, J.A, Fiedorczuk, K, Sazanov, L.A. | | Deposit date: | 2016-04-08 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | The architecture of respiratory supercomplexes.
Nature, 537, 2016
|
|
5I1S
 
 | | Villin headpiece subdomain with a Lys30 to APC substitution | | Descriptor: | D-Villin headpiece subdomain, Villin-1 | | Authors: | Kreitler, D.F, Mortenson, D.E, Gellman, S.H, Forest, K.T. | | Deposit date: | 2016-02-05 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Effects of Single alpha-to-beta Residue Replacements on Structure and Stability in a Small Protein: Insights from Quasiracemic Crystallization.
J.Am.Chem.Soc., 138, 2016
|
|
5I68
 
 | | Alcohol oxidase from Pichia pastoris | | Descriptor: | Alcohol oxidase 1, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Vonck, J, Mills, D.J, Parcej, D.N. | | Deposit date: | 2016-02-16 | | Release date: | 2016-08-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structure of Alcohol Oxidase from Pichia pastoris by Cryo-Electron Microscopy.
Plos One, 11, 2016
|
|
5I6M
 
 | | Crystal Structure of Copper Nitrite Reductase at 100K after 7.59 MGy | | Descriptor: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Horrell, S, Hough, M.A, Strange, R.W. | | Deposit date: | 2016-02-16 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Serial crystallography captures enzyme catalysis in copper nitrite reductase at atomic resolution from one crystal.
Iucrj, 3, 2016
|
|
5I9V
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with AGS | | Descriptor: | Ephrin type-A receptor 2, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.458 Å) | | Cite: | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
5IA3
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with PD173955 | | Descriptor: | 6-(2,6-DICHLORO-PHENYL)-8-METHYL-2-(3-METHYLSULFANYL-PHENYLAMINO)-8H-PYRIDO[2,3-D]PYRIMIDIN-7-ONE, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.788 Å) | | Cite: | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
5IAJ
 
 | | Caspase 3 V266L | | Descriptor: | ACE-ASP-GLU-VAL-ASK, Caspase-3, SODIUM ION | | Authors: | Maciag, J.J, Mackenzie, S.H, Tucker, M.B, Schipper, J.L, Swartz, P.D, Clark, A.C. | | Deposit date: | 2016-02-21 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Tunable allosteric library of caspase-3 identifies coupling between conserved water molecules and conformational selection.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1P2Y
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME P450CAM IN COMPLEX WITH (S)-(-)-NICOTINE | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, Cytochrome P450-cam, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Strickler, M, Goldstein, B.M, Maxfield, K, Shireman, L, Kim, G, Matteson, D, Jones, J.P. | | Deposit date: | 2003-04-16 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic Studies on the Complex Behavior of Nicotine Binding to P450cam (CYP101)(dagger).
Biochemistry, 42, 2003
|
|
8DSN
 
 | | Peptidylglycine alpha hydroxylating monoxygenase, Q272A | | Descriptor: | COPPER (II) ION, GLYCEROL, Peptidylglycine alpha-amidating monooxygenase | | Authors: | Arias, R.J, Blackburn, N.J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | New structures reveal flexible dynamics between the subdomains of peptidylglycine monooxygenase. Implications for an open to closed mechanism.
Protein Sci., 32, 2023
|
|
6B7Z
 
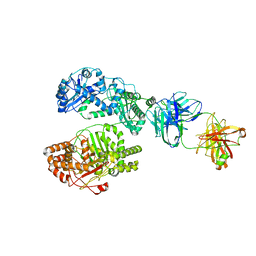 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11 heavy chain and FAB H11 light chain | | Descriptor: | FAB H11 heavy chain, FAB H11 light chain, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-05 | | Release date: | 2018-01-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
5IOU
 
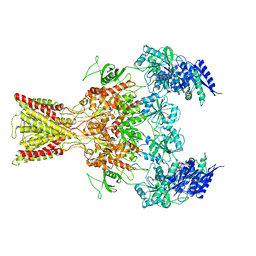 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the glutamate/glycine-bound conformation | | Descriptor: | GLUTAMIC ACID, GLYCINE, Ionotropic glutamate receptor subunit NR2B, ... | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
5IPU
 
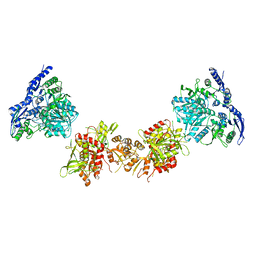 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 6 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (15.4 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
8DSL
 
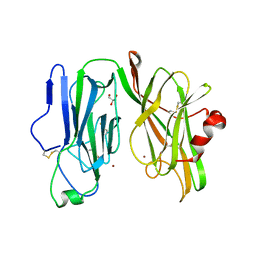 | | Peptidylglycine alpha hydroxylating monooxygenase, Q272E | | Descriptor: | COPPER (II) ION, GLYCEROL, Peptidylglycine alpha-amidating monooxygenase | | Authors: | Arias, R.J, Blackburn, N.J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | New structures reveal flexible dynamics between the subdomains of peptidylglycine monooxygenase. Implications for an open to closed mechanism.
Protein Sci., 32, 2023
|
|
1DDS
 
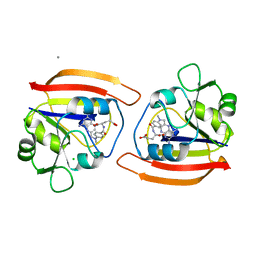 | |
1DDR
 
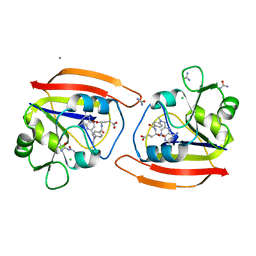 | |
1Z8L
 
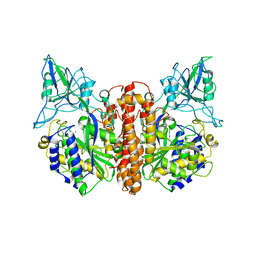 | | Crystal structure of prostate-specific membrane antigen, a tumor marker and peptidase | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Davis, M.I, Bennett, M.J, Thomas, L.M, Bjorkman, P.J. | | Deposit date: | 2005-03-30 | | Release date: | 2005-04-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of prostate-specific membrane antigen, a tumor marker and peptidase
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
5IS2
 
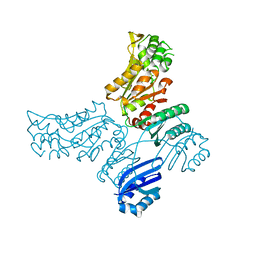 | |
5MQX
 
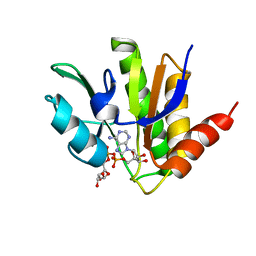 | | NMR solution structure of macro domain from Venezuelan equine encephalitis virus(VEEV) in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein3 | | Authors: | Makrynitsa, G.I, Ntonti, D, Marousis, K.D, Matsoukas, M.T, Papageorgiou, N, Coutard, B, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2016-12-21 | | Release date: | 2018-07-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational plasticity of the VEEV macro domain is important for binding of ADP-ribose.
J.Struct.Biol., 206, 2019
|
|
5MN3
 
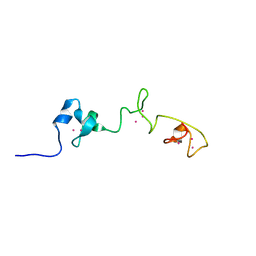 | |
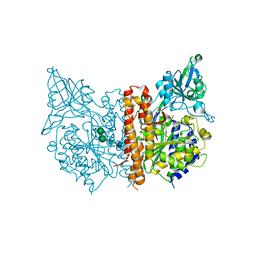
3D7D
 
 | |
5ML1
 
 | | NMR Structure of the Littorina littorea metallothionein, a snail MT folding into three distinct domains | | Descriptor: | CADMIUM ION, Putative metallothionein | | Authors: | Baumann, C, Beil, A, Jurt, S, Zerbe, O. | | Deposit date: | 2016-12-06 | | Release date: | 2017-04-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Adaptation of a Protein to Increased Metal Stress: NMR Structure of a Marine Snail Metallothionein with an Additional Domain.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MTI
 
 | | Bamb_5917 Acyl-Carrier Protein | | Descriptor: | Phosphopantetheine-binding protein | | Authors: | Gallo, A, Kosol, S, Griffiths, D, Masschelein, J, Alkhalaf, L, Smith, H, Valentic, T, Tsai, S, Challis, G, Lewandowski, J.R. | | Deposit date: | 2017-01-09 | | Release date: | 2018-08-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for chain release from the enacyloxin polyketide synthase
Nat.Chem., 2019
|
|
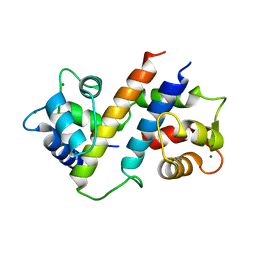
6B8Q
 
 | |
5JD0
 
 | | crystal structure of ARAP3 RhoGAP domain | | Descriptor: | Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3 | | Authors: | Bao, H, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2016-04-15 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Specific Recognition of RhoA by the Dual GTPase-activating Protein ARAP3
J.Biol.Chem., 291, 2016
|
|
5JJ8
 
 | | Crystal Structure of the Beta Carbonic Anhydrase psCA3 isolated from Pseudomonas aeruginosa - alternate crystal packing form | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Pinard, M.A, Kurian, J.J, Aggarwal, M, Agbandje-McKenna, M, McKenna, R. | | Deposit date: | 2016-04-22 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.585 Å) | | Cite: | Cryoannealing-induced space-group transition of crystals of the carbonic anhydrase psCA3.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
