2QXL
 
 | | Crystal Structure Analysis of Sse1, a yeast Hsp110 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein homolog SSE1, MAGNESIUM ION, ... | | Authors: | Hendrickson, W.A, Liu, Q. | | Deposit date: | 2007-08-12 | | Release date: | 2007-10-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Insights into hsp70 chaperone activity from a crystal structure of the yeast hsp110 Sse1.
Cell(Cambridge,Mass.), 131, 2007
|
|
2QZV
 
 | | Draft Crystal Structure of the Vault Shell at 9 Angstroms Resolution | | Descriptor: | Major vault protein | | Authors: | Anderson, D.H, Kickhoefer, V.A, Sievers, S.A, Rome, L.H, Eisenberg, D. | | Deposit date: | 2007-08-17 | | Release date: | 2007-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (9 Å) | | Cite: | Draft crystal structure of the vault shell at 9-A resolution.
Plos Biol., 5, 2007
|
|
2R1Q
 
 | |
2QKD
 
 | | Crystal structure of tandem ZPR1 domains | | Descriptor: | FORMIC ACID, ZINC ION, Zinc finger protein ZPR1 | | Authors: | Mishra, A.K. | | Deposit date: | 2007-07-10 | | Release date: | 2007-08-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the interaction of the evolutionarily conserved ZPR1 domain tandem with eukaryotic EF1A, receptors, and SMN complexes.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2QJH
 
 | |
2OWN
 
 | |
2P1W
 
 | |
2PAH
 
 | |
2PB7
 
 | |
2PBI
 
 | | The multifunctional nature of Gbeta5/RGS9 revealed from its crystal structure | | Descriptor: | GLYCEROL, Guanine nucleotide-binding protein subunit beta 5, Regulator of G-protein signaling 9 | | Authors: | Cheever, M.L, Snyder, J.T, Gershburg, S, Siderovski, D.P, Harden, T.K, Sondek, J. | | Deposit date: | 2007-03-28 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the multifunctional Gbeta5-RGS9 complex.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2OS7
 
 | | Caf1M periplasmic chaperone tetramer | | Descriptor: | Chaperone protein caf1M | | Authors: | Knight, S.D, Zavialov, A.Z. | | Deposit date: | 2007-02-05 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A novel self-capping mechanism controls aggregation of periplasmic chaperone Caf1M
MOL.MICROBIOL., 64, 2007
|
|
2P6T
 
 | | CRYSTAL STRUCTURE OF TRANSCRIPTIONAL REGULATOR NMB0573 and L-LEUCINE COMPLEX FROM NEISSERIA MENINGITIDIS | | Descriptor: | CALCIUM ION, GLYCEROL, LEUCINE, ... | | Authors: | Ren, J, Sainsbury, S, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-03-19 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure and Transcriptional Analysis of a Global Regulator from Neisseria meningitidis.
J.Biol.Chem., 282, 2007
|
|
2OX4
 
 | | Crystal structure of putative dehydratase from Zymomonas mobilis ZM4 | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Freeman, J.C, Bain, K, Gheyi, T, Wasserman, S.R, Smith, D, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-19 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Putative Dehydratase from Zymomonas Mobilis Zm4
To be Published
|
|
2OZ3
 
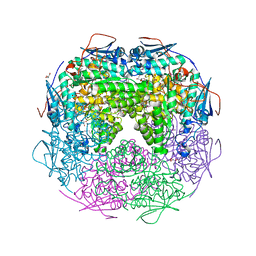 | | Crystal structure of L-Rhamnonate dehydratase from Azotobacter vinelandii | | Descriptor: | GLYCEROL, Mandelate racemase/muconate lactonizing enzyme, SODIUM ION | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Freeman, J.C, Bain, K, Gheyi, T, Wu, B, Wasserman, S.R, Smith, D, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-23 | | Release date: | 2007-03-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of L-Rhamnonate dehydratase from azotobacter vinelandii
To be Published
|
|
2P1M
 
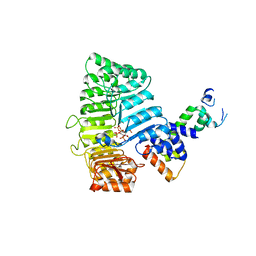 | | TIR1-ASK1 complex structure | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, SKP1-like protein 1A, TRANSPORT INHIBITOR RESPONSE 1 protein | | Authors: | Tan, X, Calderon-Villalobos, L.I.A, Sharon, M, Robinson, C.V, Estelle, M, Zheng, N. | | Deposit date: | 2007-03-05 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of auxin perception by the TIR1 ubiquitin ligase
Nature, 446, 2007
|
|
2P8R
 
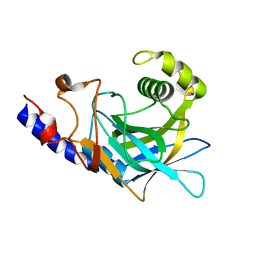 | | Crystal structure of the C-terminal domain of C. elegans pre-mRNA splicing factor Prp8 carrying R2303K mutant | | Descriptor: | Pre-mRNA-splicing factor Prp8 | | Authors: | Zhang, L, Shen, J, Guarnieri, M.T, Heroux, A, Yang, K, Zhao, R. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the C-terminal domain of splicing factor Prp8 carrying retinitis pigmentosa mutants
Protein Sci., 16, 2007
|
|
2PBW
 
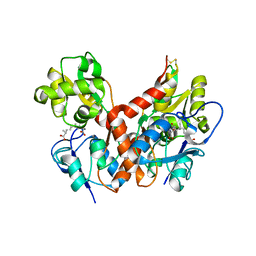 | | Crystal Structure of the Ligand-Binding Core of iGluR5 in Complex with the Partial agonist Domoic Acid at 2.5 A Resolution | | Descriptor: | (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID, Glutamate receptor, ionotropic kainate 1 | | Authors: | Hald, H, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-03-29 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Partial agonism and antagonism of the ionotropic glutamate receptor iGLuR5: structures of the ligand-binding core in complex with domoic acid and 2-amino-3-[5-tert-butyl-3-(phosphonomethoxy)-4-isoxazolyl]propionic acid.
J.Biol.Chem., 282, 2007
|
|
2QNN
 
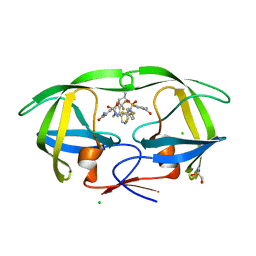 | | HIV-1 protease in complex with a multiple decorated pyrrolidine-based inhibitor | | Descriptor: | 4,4'-[(3S,4S)-pyrrolidine-3,4-diylbis({[4-(trifluoromethyl)benzyl]imino}sulfonyl)]dibenzamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2007-07-19 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure-Guided Design of C2-Symmetric HIV-1 Protease Inhibitors Based on a Pyrrolidine Scaffold.
J.Med.Chem., 51, 2008
|
|
2R3X
 
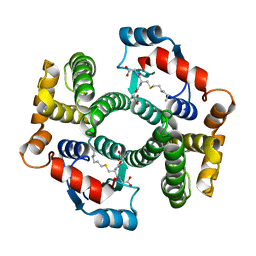 | | Crystal structure of an R15L hGSTA1-1 mutant complexed with S-hexyl-glutathione | | Descriptor: | Glutathione S-transferase A1, S-HEXYLGLUTATHIONE | | Authors: | Burke, J.P.W.G, Kinsley, N, Sayed, M, Sewell, T, Dirr, H.W. | | Deposit date: | 2007-08-30 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Arginine 15 stabilizes an S(N)Ar reaction transition state and the binding of anionic ligands at the active site of human glutathione transferase A1-1.
Biophys.Chem., 146, 2010
|
|
2QE7
 
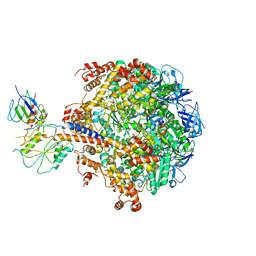 | | Crystal structure of the f1-atpase from the thermoalkaliphilic bacterium bacillus sp. ta2.a1 | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit epsilon, ... | | Authors: | Stocker, A, Keis, S, Vonck, J, Cook, G.M, Dimroth, P. | | Deposit date: | 2007-06-25 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | The Structural Basis for Unidirectional Rotation of Thermoalkaliphilic F(1)-ATPase.
Structure, 15, 2007
|
|
2QHZ
 
 | | Crystal structure of protease inhibitor, MIT-1-AC87 in complex with wild type HIV-1 protease | | Descriptor: | (2E)-N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{(2-THIENYLMETHYL)[(2,4,5-TRIFLUOROPHENYL)SULFONYL]AMINO}PROPYL]-4,4,4-TRIFLUORO-3-METHYLBUT-2-ENAMIDE, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
2QHY
 
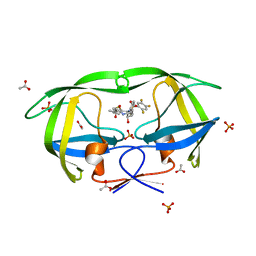 | | Crystal Structure of protease inhibitor, MIT-1-AC86 in complex with wild type HIV-1 protease | | Descriptor: | ACETATE ION, N~2~-ACETYL-N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{(2-THIENYLMETHYL)[(2,4,5-TRIFLUOROPHENYL)SULFONYL]AMINO}PROPYL]-L-ALANINAMIDE, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
2QVZ
 
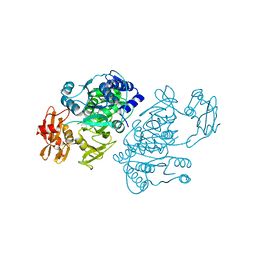 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, I303A mutation, bound to 3-Chlorobenzoate | | Descriptor: | 3-chlorobenzoate, 4-Chlorobenzoate CoA Ligase/Synthetase | | Authors: | Wu, R, Reger, A.S, Cao, J, Gulick, A.M, Dunaway-Mariano, D. | | Deposit date: | 2007-08-09 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational redesign of the 4-chlorobenzoate binding site of 4-chlorobenzoate: coenzyme a ligase for expanded substrate range.
Biochemistry, 46, 2007
|
|
2QIK
 
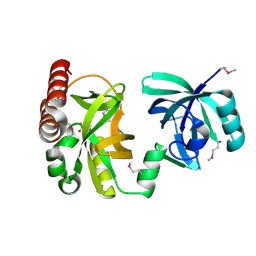 | | Crystal structure of YkqA from Bacillus subtilis. Northeast Structural Genomics Target SR631 | | Descriptor: | CITRIC ACID, UPF0131 protein ykqA | | Authors: | Benach, J, Chen, Y, Forouhar, F, Seetharaman, J, Baran, M.C, Cunningham, K, Ma, L.-C, Owens, L, Chen, C.X, Rong, X, Janjua, H, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-07-05 | | Release date: | 2007-07-24 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of YkqA from Bacillus subtilis.
To be Published
|
|
2QJY
 
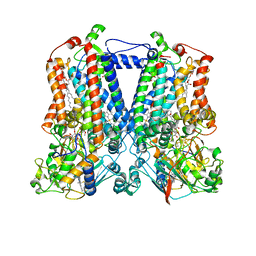 | | Crystal structure of rhodobacter sphaeroides double mutant with stigmatellin and UQ2 | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, 2-O-octyl-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Esser, L, Xia, D. | | Deposit date: | 2007-07-09 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibitor-complexed Structures of the Cytochrome bc1 from the Photosynthetic Bacterium Rhodobacter sphaeroides.
J.Biol.Chem., 283, 2008
|
|
