7RXF
 
 | |
5Z6J
 
 | |
7CSQ
 
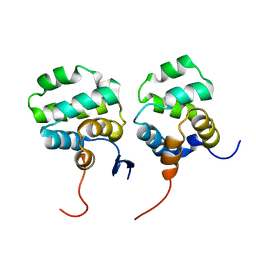 | | Solution structure of the complex between p75NTR-DD and TRADD-DD | | Descriptor: | Tumor necrosis factor receptor superfamily member 16, Tumor necrosis factor receptor type 1-associated DEATH domain protein | | Authors: | Lin, Z, Zhang, N. | | Deposit date: | 2020-08-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of NF-kappa B signaling by the p75 neurotrophin receptor interaction with adaptor protein TRADD through their respective death domains.
J.Biol.Chem., 297, 2021
|
|
5FCK
 
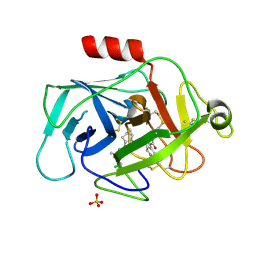 | | COMPLEMENT FACTOR D IN COMPLEX WITH COMPOUND 5 | | Descriptor: | 1-[2-[(1~{R},3~{S},5~{R})-3-[[(1~{R})-1-(3-chloranyl-2-fluoranyl-phenyl)ethyl]carbamoyl]-2-azabicyclo[3.1.0]hexan-2-yl]-2-oxidanylidene-ethyl]pyrazolo[3,4-c]pyridine-3-carboxamide, Complement factor D, SULFATE ION | | Authors: | Mac Sweeney, A. | | Deposit date: | 2015-12-15 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
5FDA
 
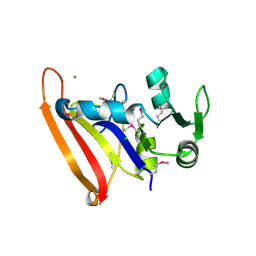 | | The high resolution structure of apo form dihydrofolate reductase from Yersinia pestis at 1.55 A | | Descriptor: | CHLORIDE ION, Dihydrofolate reductase | | Authors: | Chang, C, Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-15 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | structure of dihydrofolate reductase from Yersinia pestis complex with
To Be Published
|
|
3BI1
 
 | |
3B5W
 
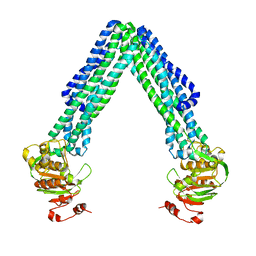 | | Crystal Structure of Eschericia coli MsbA | | Descriptor: | Lipid A export ATP-binding/permease protein msbA | | Authors: | Ward, A, Reyes, C.L, Yu, J, Roth, C.B, Chang, G. | | Deposit date: | 2007-10-26 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (5.3 Å) | | Cite: | Flexibility in the ABC transporter MsbA: Alternating access with a twist.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1Q5C
 
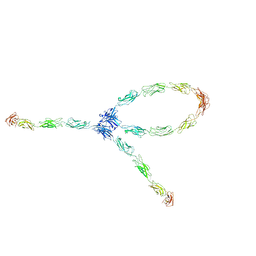 | | S-S-lambda-shaped TRANS and CIS interactions of cadherins model based on fitting C-cadherin (1L3W) to 3D map of desmosomes obtained by electron tomography | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | He, W, Cowin, P, Stokes, D.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (30 Å) | | Cite: | Untangling Desmosomal Knots with Electron Tomography
Science, 302, 2003
|
|
5D6K
 
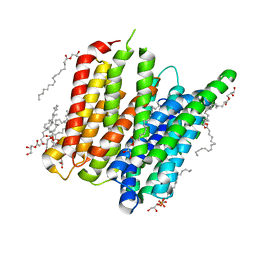 | | PepT - CIM | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, Di-or tripeptide:H+ symporter, ... | | Authors: | Ma, P, Caffrey, M. | | Deposit date: | 2015-08-12 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The cubicon method for concentrating membrane proteins in the cubic mesophase.
Nat Protoc, 12, 2017
|
|
3ZIE
 
 | | SepF-like protein from Archaeoglobus fulgidus | | Descriptor: | SEPF-LIKE PROTEIN | | Authors: | Duman, R, Ishikawa, S, Celik, I, Ogasawara, N, Lowe, J, Hamoen, L.W. | | Deposit date: | 2013-01-08 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Genetic Analyses Reveal the Protein Sepf as a New Membrane Anchor for the Z Ring
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3BHX
 
 | |
1Q5B
 
 | | lambda-shaped TRANS and CIS interactions of cadherins model based on fitting C-cadherin (1L3W) to 3D map of desmosomes obtained by electron tomography | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | He, W, Cowin, P, Stokes, D.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (30 Å) | | Cite: | Untangling Desmosomal Knots with Electron Tomography
Science, 302, 2003
|
|
5H1I
 
 | | NMR structure of TIBA, a chimera of SFTI | | Descriptor: | Bradykinin-trypsin inhibitor secondary loop chimera | | Authors: | Xiao, T, Tam, J.P. | | Deposit date: | 2016-10-10 | | Release date: | 2017-04-19 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | An Orally Active Bradykinin B1 Receptor Antagonist Engineered as a Bifunctional Chimera of Sunflower Trypsin Inhibitor.
J. Med. Chem., 60, 2017
|
|
5GIW
 
 | | Solution NMR structure of Humanin containing a D-isomerized serine residue | | Descriptor: | Humanin | | Authors: | Furuita, K, Sugiki, T, Alsanousi, N, Fujiwara, T, Kojima, C. | | Deposit date: | 2016-06-25 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure and inhibitory effect against amyloid-beta fibrillation of Humanin containing a d-isomerized serine residue
Biochem.Biophys.Res.Commun., 477, 2016
|
|
1KO1
 
 | | Crystal structure of gluconate kinase | | Descriptor: | CHLORIDE ION, Gluconate kinase | | Authors: | Kraft, L, Sprenger, G.A, Lindqvist, Y. | | Deposit date: | 2001-12-20 | | Release date: | 2002-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Conformational changes during the catalytic cycle of gluconate kinase as revealed by X-ray crystallography.
J.Mol.Biol., 318, 2002
|
|
5H3M
 
 | |
5G1X
 
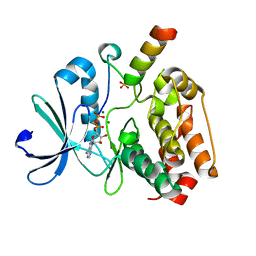 | | Crystal structure of Aurora-A kinase in complex with N-Myc | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AURORA KINASE A, MAGNESIUM ION, ... | | Authors: | Richards, M.W, Burgess, S.G, Bayliss, R. | | Deposit date: | 2016-03-31 | | Release date: | 2016-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis of N-Myc Binding by Aurora-A and its Destabilization by Kinase Inhibitors
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1KEQ
 
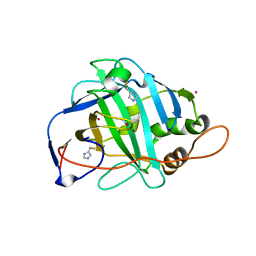 | | Crystal Structure of F65A/Y131C Carbonic Anhydrase V, covalently modified with 4-chloromethylimidazole | | Descriptor: | 4-METHYLIMIDAZOLE, ACETIC ACID, F65A/Y131C-MI Carbonic Anhydrase V, ... | | Authors: | Jude, K.M, Wright, S.K, Tu, C, Silverman, D.N, Viola, R.E, Christianson, D.W. | | Deposit date: | 2001-11-16 | | Release date: | 2002-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of F65A/Y131C-methylimidazole carbonic anhydrase V reveals architectural features of an engineered proton shuttle.
Biochemistry, 41, 2002
|
|
4RHV
 
 | |
5AND
 
 | | Crystal structure of CDK2 in complex with 2-imidazol-1-yl-1H- benzimidazole processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | 2-IMIDAZOL-1-YL-1H-BENZIMIDAZOLE, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Zander, U, Hoffmann, G, Mathieu, M, Marquette, J.-P, Cornaciu, I, Cipriani, F, Marquez, J.A. | | Deposit date: | 2015-09-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Automated Harvesting and Processing of Protein Crystals Through Laser Photoablation.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5ANE
 
 | | Crystal structure of CDK2 in complex with 6-methoxy-7H-purine processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | 6-METHOXY-9H-PURINE, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Zander, U, Hoffmann, G, Mathieu, M, Marquette, J.-P, Cornaciu, I, Cipriani, F, Marquez, J.A. | | Deposit date: | 2015-09-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Automated Harvesting and Processing of Protein Crystals Through Laser Photoablation.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
4HZM
 
 | | Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) bound to N-[(3S,4R,5R,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]butanamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-hexosaminidase, N-[(3S,4R,5R,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]butanamide | | Authors: | Bacik, J.P, Mark, B.L. | | Deposit date: | 2012-11-15 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Development of Selective Inhibitors of NagZ: Increased Susceptibility of Gram-Negative Bacteria to beta-Lactams.
Chembiochem, 14, 2013
|
|
5ANJ
 
 | | Crystal structure of CDK2 in complex with N-(9H-purin-6-yl)thiophene- 2-carboxamide processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | CYCLIN-DEPENDENT KINASE 2, N-(9H-purin-6-yl)thiophene-2-carboxamide | | Authors: | Zander, U, Hoffmann, G, Mathieu, M, Marquette, J.-P, Cornaciu, I, Cipriani, F, Marquez, J.A. | | Deposit date: | 2015-09-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Automated Harvesting and Processing of Protein Crystals Through Laser Photoablation.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5AMW
 
 | | Crystal Structure of the Strawberry Pathogenesis-Related 10 (PR-10) Fra a 2 protein (A141F) processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | Fra a 2 allergen | | Authors: | Zander, U, Casanal, A, Pott, D, Valpuesta, V, Hoffmann, G, Cornaciu, I, Cipriani, F, Marquez, J.A. | | Deposit date: | 2015-09-02 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Automated Harvesting and Processing of Protein Crystals Through Laser Photoablation.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5ANG
 
 | | Crystal structure of CDK2 in complex with 7-hydroxy-4-(morpholinomethyl)chromen-2-one processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | 7-HYDROXY-4-(MORPHOLINOMETHYL)CHROMEN-2-ONE, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Zander, U, Hoffmann, G, Mathieu, M, Marquette, J.-P, Cornaciu, I, Cipriani, F, Marquez, J.A. | | Deposit date: | 2015-09-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Automated Harvesting and Processing of Protein Crystals Through Laser Photoablation.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
