3LBC
 
 | |
3LCW
 
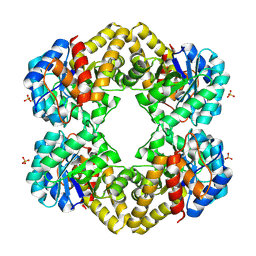 | | L-KDO aldolase complexed with hydroxypyruvate | | Descriptor: | 3-HYDROXYPYRUVIC ACID, N-acetylneuraminate lyase, SULFATE ION | | Authors: | Chou, C.-Y, Wang, A.H.-J, Ko, T.-P. | | Deposit date: | 2010-01-12 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Modulation of substrate specificities of D-sialic acid aldolase through single mutations of Val251
To be Published
|
|
3M5V
 
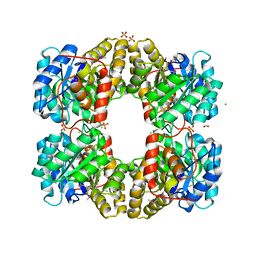 | | Crystal Structure of Dihydrodipicolinate Synthase from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Dihydrodipicolinate synthase, ... | | Authors: | Kim, Y, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-13 | | Release date: | 2010-04-28 | | Last modified: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Dihydrodipicolinate Synthase from Campylobacter jejuni
To be Published
|
|
3N2X
 
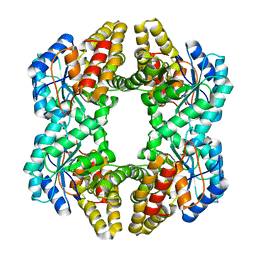 | | Crystal structure of YagE, a prophage protein belonging to the dihydrodipicolinic acid synthase family from E. coli K12 in complex with pyruvate | | Descriptor: | 1,2-ETHANEDIOL, Uncharacterized protein yagE | | Authors: | Bhaskar, V, Kumar, P.M, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2010-05-19 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of biochemical and putative biological role of a xenolog from Escherichia coli using structural analysis.
Proteins, 79, 2011
|
|
3NEV
 
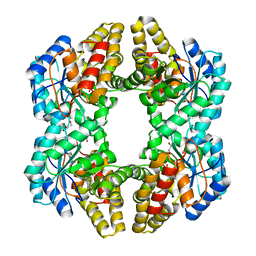 | | Crystal structure of YagE, a prophage protein from E. coli K12 in complex with KDGal | | Descriptor: | 1,2-ETHANEDIOL, 3-DEOXY-D-LYXO-HEXONIC ACID, Uncharacterized protein yagE | | Authors: | Bhaskar, V, Kumar, P.M, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2010-06-09 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identification of biochemical and putative biological role of a xenolog from Escherichia coli using structural analysis.
Proteins, 79, 2011
|
|
3NA8
 
 | | Crystal Structure of a putative dihydrodipicolinate synthetase from Pseudomonas aeruginosa | | Descriptor: | D-MALATE, MAGNESIUM ION, putative dihydrodipicolinate synthetase | | Authors: | Qiu, W, Lam, R, Romanov, V, Jones, K, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2010-06-01 | | Release date: | 2011-06-01 | | Last modified: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of a putative dihydrodipicolinate synthetase from Pseudomonas aeruginosa
To be Published
|
|
3NOE
 
 | | Crystal Structure of Dihydrodipicolinate synthase from Pseudomonas aeruginosa | | Descriptor: | Dihydrodipicolinate synthase, S-1,2-PROPANEDIOL | | Authors: | Kaur, N, Kumar, S, Singh, N, Gautam, A, Sharma, R, Sharma, S, Tewari, R, Singh, T.P. | | Deposit date: | 2010-06-25 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of Dihydrodipicolinate synthase from Pseudomonas aeruginosa
To be Published
|
|
3PB0
 
 | |
3PB2
 
 | |
7JZD
 
 | |
7JZE
 
 | |
7JZA
 
 | |
7JZC
 
 | |
7JZF
 
 | |
7JZB
 
 | |
7JZG
 
 | |
7JZ9
 
 | |
7JZ7
 
 | | Dihydrodipicolinate synthase mutant S48F | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, GLYCEROL, POTASSIUM ION | | Authors: | Board, A.J, Dobson, R.C.J. | | Deposit date: | 2020-09-01 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Mapping the unchartered waters of dihydrodipicolinate synthase: a novel mechanism of allosteric inhibition.
To Be Published
|
|
7JZ8
 
 | |
7KEL
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, H59K mutant with pyruvate bound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Sanders, D.A.R. | | Deposit date: | 2020-10-11 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reversing the roles of a crucial hydrogen-bonding pair: a lysine-insensitive mutant of Campylobacter jejuni dihydrodipicolinate synthase, H59K, binds histidine in its allosteric site
To Be Published
|
|
7KG9
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, H56W mutant with pyruvate bound in the active site and L-lysine bound at the allosteric site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Sanders, D.A.R. | | Deposit date: | 2020-10-16 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Fluorescence-based Assay Development for Screening Novel Inhibitors of Dihydrodipicolinate Synthase from Campylobacter jejuni
To Be Published
|
|
7KG5
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, H56W mutant with pyruvate bound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Sanders, D.A.R. | | Deposit date: | 2020-10-15 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fluorescence-based Assay Development for Screening Novel Inhibitors of Dihydrodipicolinate Synthase from Campylobacter jejuni
To Be Published
|
|
7KG2
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, H59K mutant with pyruvate bound in the active site and L-histidine bound at the allosteric site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Sanders, D.A.R. | | Deposit date: | 2020-10-15 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Reversing the roles of a crucial hydrogen-bonding pair: a lysine-insensitive mutant of Campylobacter jejuni dihydrodipicolinate synthase, H59K, binds histidine in its allosteric site
To be Published
|
|
7KH4
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, H56W mutant with pyruvate bound in the active site and R,R-bislysine bound at the allosteric site | | Descriptor: | (2R,5R)-2,5-diamino-2,5-bis(4-aminobutyl)hexanedioic acid, 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ... | | Authors: | Saran, S, Majdi Yazdi, M, Sanders, D.A.R. | | Deposit date: | 2020-10-19 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fluorescence-based Assay Development for Screening Novel Inhibitors of Dihydrodipicolinate Synthase from Campylobacter jejuni
To Be Published
|
|
7KK1
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, N84A mutant with pyruvate bound in the active site and L-lysine bound at the allosteric site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Sanders, D.A.R. | | Deposit date: | 2020-10-27 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A TIGHT DIMER INTERFACE N84 RESIDUE, PLAYS A CRITICAL ROLE IN THE TRANSMISSION OF THE ALLOSTERIC INHIBITION SIGNALS IN Cj.DHDPS
To Be Published
|
|
