3DY0
 
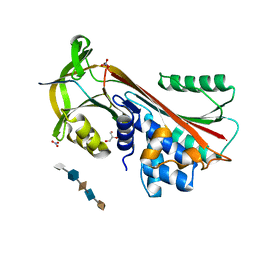 | | Crystal Structure of Cleaved PCI Bound to Heparin | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, C-terminus Plasma serine protease inhibitor, GLYCEROL, ... | | Authors: | Li, W, Huntington, J.A. | | Deposit date: | 2008-07-25 | | Release date: | 2008-10-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The heparin binding site of protein C inhibitor is protease-dependent.
J.Biol.Chem., 283, 2008
|
|
1IZ2
 
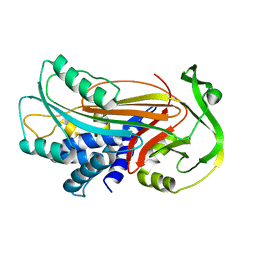 | | Interactions causing the kinetic trap in serpin protein folding | | Descriptor: | alpha-D-glucopyranose-(1-2)-(5R)-5-[(2R)-2-hydroxynonyl]-beta-D-xylulofuranose, alpha1-antitrypsin | | Authors: | Im, H, Woo, M.-S, Hwang, K.Y, Yu, M.-H. | | Deposit date: | 2002-09-19 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions causing the kinetic trap in serpin protein folding
J.BIOL.CHEM., 277, 2002
|
|
3EOX
 
 | |
1IMV
 
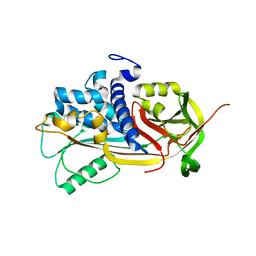 | | 2.85 A crystal structure of PEDF | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PIGMENT EPITHELIUM-DERIVED FACTOR | | Authors: | Simonovic, M, Gettins, P.G.W, Volz, K. | | Deposit date: | 2001-05-11 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of human PEDF, a potent anti-angiogenic and neurite growth-promoting factor.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JRR
 
 | | HUMAN PLASMINOGEN ACTIVATOR INHIBITOR-2.[LOOP (66-98) DELETIONMUTANT] COMPLEXED WITH PEPTIDE MIMIckING THE REACTIVE CENTER LOOP | | Descriptor: | BETA-MERCAPTOETHANOL, PLASMINOGEN ACTIVATOR INHIBITOR-2 | | Authors: | Jankova, L, Harrop, S.J, Saunders, D.N, Andrews, J.L, Bertram, K.C, Gould, A.R, Baker, M.S, Curmi, P.M.G. | | Deposit date: | 2001-08-15 | | Release date: | 2001-09-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF THE COMPLEX OF PLASMINOGEN ACTIVATOR INHIBITOR 2 WITH A PEPTIDE MIMICKING THE REACTIVE CENTER LOOP
J.Biol.Chem., 276, 2001
|
|
1JVQ
 
 | | Crystal structure at 2.6A of the ternary complex between antithrombin, a P14-P8 reactive loop peptide, and an exogenous tetrapeptide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTITHROMBIN-III, ... | | Authors: | Zhou, A, Huntington, J.A, Lomas, D.A, Carrell, R.W, Stein, P.E. | | Deposit date: | 2001-08-31 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | How small peptides block and reverse serpin polymerisation
J.Mol.Biol., 342, 2004
|
|
1JTI
 
 | |
1JJO
 
 | |
2OL2
 
 | |
5OM6
 
 | |
5OM8
 
 | |
5OM5
 
 | |
5OM2
 
 | | Crystal structure of Alpha1-antichymotrypsin variant DBS-I1: a drug-binding serpin for doxycycline | | Descriptor: | (4S,4AR,5S,5AR,6R,12AS)-4-(DIMETHYLAMINO)-3,5,10,12,12A-PENTAHYDROXY-6-METHYL-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2-CARBOXAMIDE, 1,2-ETHANEDIOL, Alpha-1-antichymotrypsin | | Authors: | Schmidt, K, Muller, Y.A. | | Deposit date: | 2017-07-28 | | Release date: | 2018-05-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Design of an allosterically modulated doxycycline and doxorubicin drug-binding protein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5OM7
 
 | |
5OM3
 
 | | Crystal structure of Alpha1-antichymotrypsin variant DBS-I5: a MMP14-cleavable drug-binding serpin for doxycycline | | Descriptor: | (4S,4AR,5S,5AR,6R,12AS)-4-(DIMETHYLAMINO)-3,5,10,12,12A-PENTAHYDROXY-6-METHYL-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2-CARBOXAMIDE, Alpha-1-antichymotrypsin, DI(HYDROXYETHYL)ETHER | | Authors: | Schmidt, K, Muller, Y.A. | | Deposit date: | 2017-07-28 | | Release date: | 2018-05-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of an allosterically modulated doxycycline and doxorubicin drug-binding protein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2QUG
 
 | | Crystal structure of alpha-1-antitrypsin, crystal form A | | Descriptor: | Alpha-1-antitrypsin | | Authors: | Hansen, G, Morton, C.J, Pearce, M.C, Feil, S.C, Adams, J.J, Parker, M.W, Bottomley, S.P. | | Deposit date: | 2007-08-05 | | Release date: | 2008-08-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Preventing serpin aggregation: The molecular mechanism of citrate action upon antitrypsin unfolding.
Protein Sci., 17, 2008
|
|
2PEE
 
 | | Crystal Structure of a Thermophilic Serpin, Tengpin, in the Native State | | Descriptor: | GLYCEROL, SULFATE ION, Serine protease inhibitor | | Authors: | Zhang, Q.W, Buckle, A.M, Whisstock, J.C. | | Deposit date: | 2007-04-02 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The N terminus of the serpin, tengpin, functions to trap the metastable native state.
Embo Rep., 8, 2007
|
|
2R9Y
 
 | | Structure of antiplasmin | | Descriptor: | Alpha-2-antiplasmin | | Authors: | Law, R.H.P, Sofian, T, Kan, W.T, Horvath, A.J, Hitchen, C.R, Langendorf, C.G, Buckle, A.M, Whisstock, J.C, Coughlin, P.B. | | Deposit date: | 2007-09-14 | | Release date: | 2007-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | X-ray crystal structure of the fibrinolysis inhibitor {alpha}2-antiplasmin
Blood, 111, 2008
|
|
5NCW
 
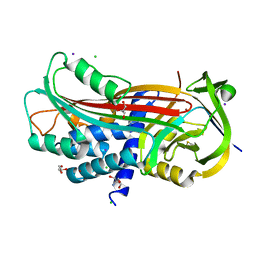 | | Structure of the trypsin induced serpin-type proteinase inhibitor, miropin (V367K/K368A mutant). | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Goulas, T, Ksiazek, M, Garcia-Ferrer, I, Mizgalska, D, Potempa, J, Gomis-Ruth, X. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
2RIV
 
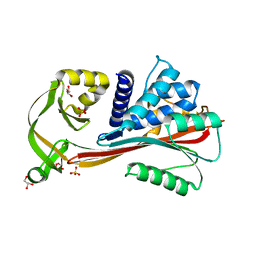 | | Crystal structure of the reactive loop cleaved human Thyroxine Binding Globulin | | Descriptor: | GLYCEROL, SULFATE ION, Thyroxine-binding globulin | | Authors: | Zhou, A, Wei, Z, Stanley, P.L.D, Read, R.J, Stein, P.E, Carrell, R.W. | | Deposit date: | 2007-10-12 | | Release date: | 2008-10-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Allosteric modulation of hormone release from thyroxine and corticosteroid-binding globulins
J.Biol.Chem., 286, 2011
|
|
5NBU
 
 | |
2RIW
 
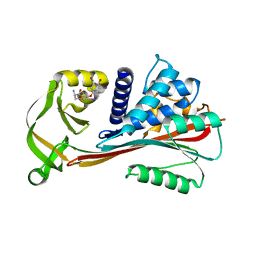 | | The Reactive loop cleaved human Thyroxine Binding Globulin complexed with thyroxine | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, Thyroxine-binding globulin | | Authors: | Zhou, A, Wei, Z, Stanley, P.L.D, Read, R.J, Stein, P.E, Carrell, R.W. | | Deposit date: | 2007-10-12 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Allosteric modulation of hormone release from thyroxine and corticosteroid-binding globulins
J.Biol.Chem., 286, 2011
|
|
5NCS
 
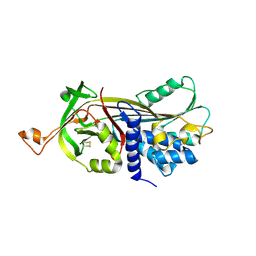 | | Structure of the native serpin-type proteinase inhibitor, miropin. | | Descriptor: | Serpin | | Authors: | Goulas, T, Ksiazek, M, Garcia-Ferrer, I, Mizgalska, D, Potempa, J, Gomis-Ruth, X. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
5NCU
 
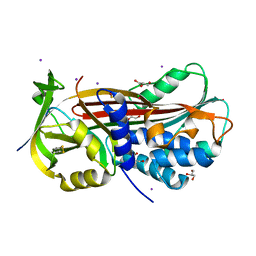 | | Structure of the subtilisin induced serpin-type proteinase inhibitor, miropin. | | Descriptor: | CHLORIDE ION, GLYCEROL, IODIDE ION, ... | | Authors: | Goulas, T, Ksiazek, M, Garcia-Ferrer, I, Mizgalska, D, Potempa, J, Gomis-Ruth, X. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
5NCT
 
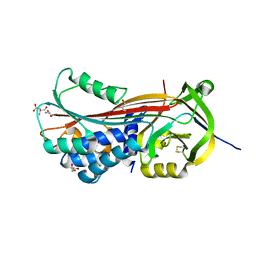 | | Structure of the trypsin induced serpin-type proteinase inhibitor, miropin. | | Descriptor: | ASPARTIC ACID, GLYCEROL, SERINE, ... | | Authors: | Goulas, T, Ksiazek, M, Garcia-Ferrer, I, Mizgalska, D, Potempa, J, Gomis-Ruth, X. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
