8G6L
 
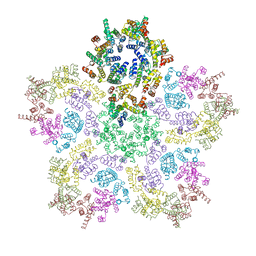 | | HIV-1 capsid lattice bound to IP6, pH 6.2 | | Descriptor: | Capsid protein | | Authors: | Highland, C.M, Dick, R.A. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into HIV-1 polyanion-dependent capsid lattice formation revealed by single particle cryo-EM.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8TY6
 
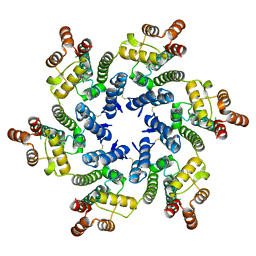 | |
8D3B
 
 | |
8GDV
 
 | |
7RJ4
 
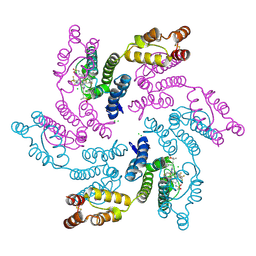 | |
8CL1
 
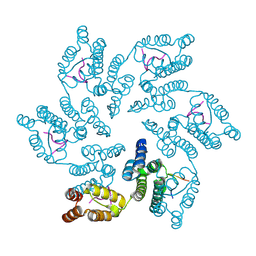 | |
6ECN
 
 | | HIV-1 CA 1/2-hexamer-EE | | Descriptor: | HIV-1 CA | | Authors: | Summers, B.J, Xiong, Y. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms.
Cell Host Microbe, 26, 2019
|
|
6EC2
 
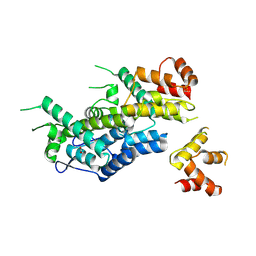 | | Structure of HIV-1 CA 1/3-hexamer | | Descriptor: | ACETATE ION, Capsid protein p24 | | Authors: | Summers, B.J, Xiong, Y. | | Deposit date: | 2018-08-07 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms.
Cell Host Microbe, 26, 2019
|
|
7URN
 
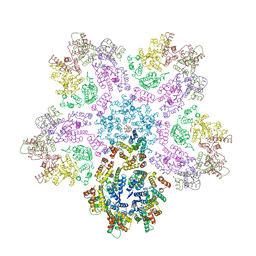 | |
8CL2
 
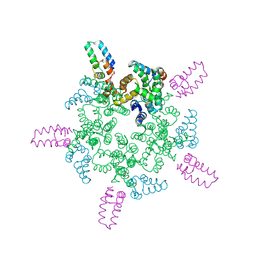 | |
7N9V
 
 | |
8G6N
 
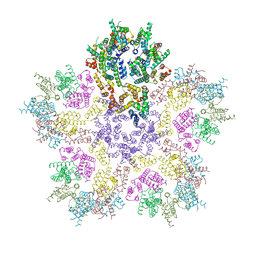 | | HIV-1 capsid lattice bound to dNTPs | | Descriptor: | Capsid protein | | Authors: | Highland, C.M, Dick, R.A. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into HIV-1 polyanion-dependent capsid lattice formation revealed by single particle cryo-EM.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7N9X
 
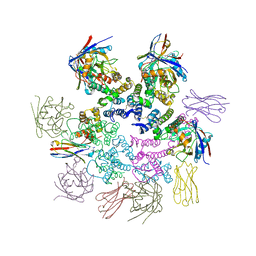 | | CA-targeting nanobody is a tool for studying HIV-1 capsid lattice interactions | | Descriptor: | Capsid protein, Nanobody, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Gerber, E.E, Digianantonio, K.M, Tripler, T.N, Smaga, S.S, Summers, B.J, Xiong, Y. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.511 Å) | | Cite: | CA-targeting nanobody is a tool for studying HIV-1 capsid lattice interactions
To Be Published
|
|
8G6K
 
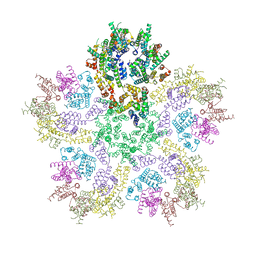 | |
8EJL
 
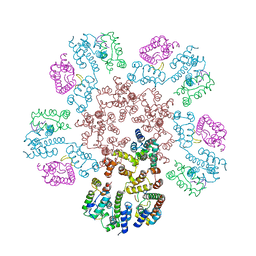 | | Structure of HIV-1 capsid declination in complex with CPSF6-FG peptide | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 6, HIV-1 capsid protein | | Authors: | Pornillos, O, Ganser-Pornillos, B.K, Schirra, R.T, dos Santos, N.F.B. | | Deposit date: | 2022-09-17 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A molecular switch modulates assembly and host factor binding of the HIV-1 capsid.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5L93
 
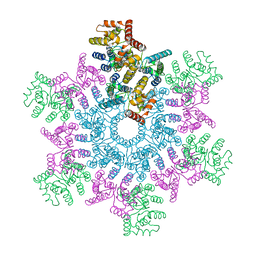 | | An atomic model of HIV-1 CA-SP1 reveals structures regulating assembly and maturation | | Descriptor: | Capsid protein p24 | | Authors: | Schur, F.K.M, Obr, M, Hagen, W.J.H, Wan, W, Arjen, J.J, Kirkpatrick, J.M, Sachse, C, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-06-09 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation.
Science, 353, 2016
|
|
6ECO
 
 | | Hexamer-2-Foldon HIV-1 capsid platform | | Descriptor: | HIV-1 capsid platform protein | | Authors: | Summers, B.J, Xiong, Y. | | Deposit date: | 2018-08-08 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms.
Cell Host Microbe, 26, 2019
|
|
5UPW
 
 | |
6VWS
 
 | | Hexamer of Helical HIV capsid by RASTR method | | Descriptor: | HIV capsid protein | | Authors: | Zhao, H, Iqbal, N, Asturias, F, Kvaratskhelia, M, Vanblerkom, P. | | Deposit date: | 2020-02-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.08 Å) | | Cite: | Structural and mechanistic bases for a potent HIV-1 capsid inhibitor.
Science, 370, 2020
|
|
5MCX
 
 | | The structure of the mature HIV-1 CA hexamer in intact virus particles | | Descriptor: | Capsid protein p24 | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
7OVR
 
 | | Mature HIV-1 matrix structure | | Descriptor: | HIV-1 matrix, MYRISTIC ACID, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Qu, K, Ke, Z.L, Zila, V, Anders-Oesswein, M, Glass, B, Muecksch, F, Mueller, R, Schultz, C, Mueller, B, Kraeusslich, H.G, Briggs, J.A.G. | | Deposit date: | 2021-06-15 | | Release date: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Maturation of the matrix and viral membrane of HIV-1.
Science, 373, 2021
|
|
3GV2
 
 | | X-ray Structure of Hexameric HIV-1 CA | | Descriptor: | Capsid protein p24,Carbon dioxide-concentrating mechanism protein CcmK homolog 4 | | Authors: | Kelly, B.N. | | Deposit date: | 2009-03-30 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | X-ray structures of the hexameric building block of the HIV capsid.
Cell(Cambridge,Mass.), 137, 2009
|
|
7OVQ
 
 | | Immature HIV-1 matrix structure | | Descriptor: | Gag polyprotein, MYRISTIC ACID | | Authors: | Qu, K, Ke, Z.L, Zila, V, Anders-Oesswein, M, Glass, B, Muecksch, F, Mueller, R, Schultz, C, Mueller, B, Kraeusslich, H.G, Briggs, J.A.G. | | Deposit date: | 2021-06-15 | | Release date: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Maturation of the matrix and viral membrane of HIV-1.
Science, 373, 2021
|
|
5MCY
 
 | | The structure of the mature HIV-1 CA pentamer in intact virus particles | | Descriptor: | Capsid protein p24 | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5UP4
 
 | |
